|
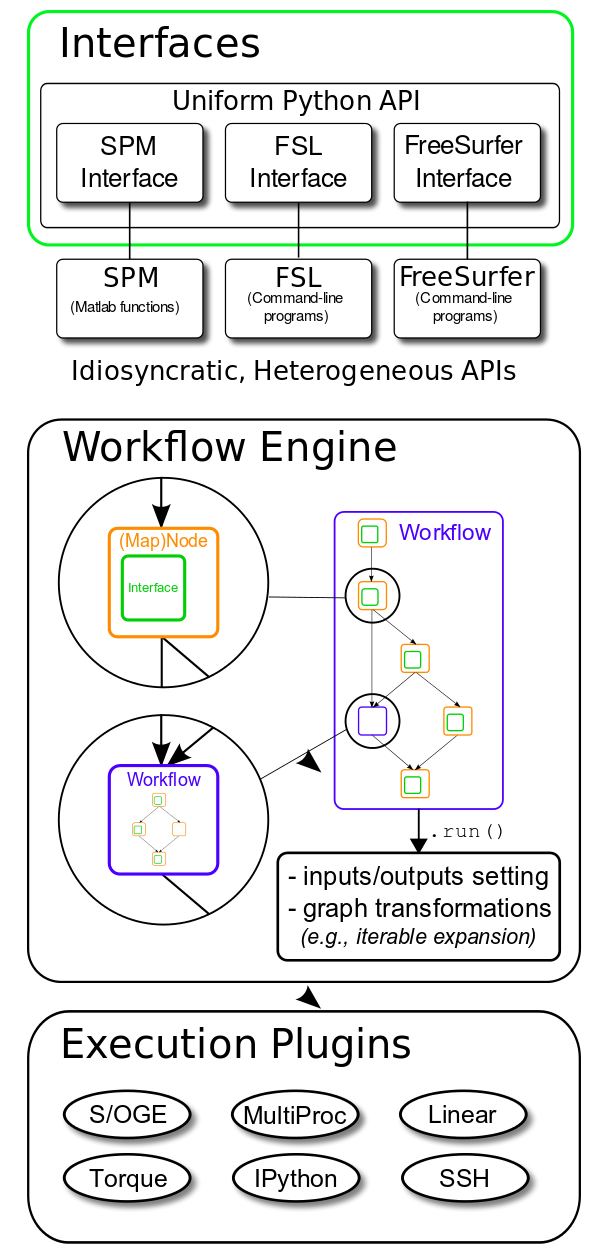
Current neuroimaging software offer users an incredible opportunity to analyze data using a variety of different algorithms. However, this has resulted in a heterogeneous collection of specialized applications without transparent interoperability or a uniform operating interface. Nipype, an open-source, community-developed initiative under the umbrella of NiPy, is a Python project that provides a uniform interface to existing neuroimaging software and facilitates interaction between these packages within a single workflow. Nipype provides an environment that encourages interactive exploration of algorithms from different packages (e.g., ANTS, SPM, FSL, FreeSurfer, Camino, MRtrix, MNE, AFNI, Slicer, DIPY), eases the design of workflows within and between packages, and reduces the learning curve necessary to use different packages. Nipype is creating a collaborative platform for neuroimaging software development in a high-level language and addressing limitations of existing pipeline systems. Nipype allows you to:
|
Reference
Gorgolewski K, Burns CD, Madison C, Clark D, Halchenko YO, Waskom ML, Ghosh SS. (2011). Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in Python. Front. Neuroinform. 5:13. Download
Tip
To get started, click Quickstart above. The Links box on the right is available on any page of this website.



