interfaces.afni.preprocess¶
Allineate¶
Wraps command 3dAllineate
Program to align one dataset (the ‘source’) to a base dataset
For complete details, see the 3dAllineate Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> allineate = afni.Allineate()
>>> allineate.inputs.in_file = 'functional.nii'
>>> allineate.inputs.out_file= 'functional_allineate.nii'
>>> allineate.inputs.in_matrix= 'cmatrix.mat'
>>> res = allineate.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dAllineate
flag: -source %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
autobox: (a boolean)
Expand the -automask function to enclose a rectangular
box that holds the irregular mask.
flag: -autobox
automask: (an integer (int or long))
Compute a mask function, set a value for dilation or 0.
flag: -automask+%d
autoweight: (a string)
Compute a weight function using the 3dAutomask
algorithm plus some blurring of the base image.
flag: -autoweight%s
center_of_mass: (a string)
Use the center-of-mass calculation to bracket the shifts.
flag: -cmass%s
check: (a list of items which are 'leastsq' or 'ls' or 'mutualinfo'
or 'mi' or 'corratio_mul' or 'crM' or 'norm_mutualinfo' or 'nmi' or
'hellinger' or 'hel' or 'corratio_add' or 'crA' or 'corratio_uns'
or 'crU')
After cost functional optimization is done, start at the
final parameters and RE-optimize using this new cost functions.
If the results are too different, a warning message will be
printed. However, the final parameters from the original
optimization will be used to create the output dataset.
flag: -check %s
convergence: (a float)
Convergence test in millimeters (default 0.05mm).
flag: -conv %f
cost: ('leastsq' or 'ls' or 'mutualinfo' or 'mi' or 'corratio_mul' or
'crM' or 'norm_mutualinfo' or 'nmi' or 'hellinger' or 'hel' or
'corratio_add' or 'crA' or 'corratio_uns' or 'crU')
Defines the 'cost' function that defines the matching
between the source and the base
flag: -cost %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
epi: (a boolean)
Treat the source dataset as being composed of warped
EPI slices, and the base as comprising anatomically
'true' images. Only phase-encoding direction image
shearing and scaling will be allowed with this option.
flag: -EPI
final_interpolation: ('nearestneighbour' or 'linear' or 'cubic' or
'quintic' or 'wsinc5')
Defines interpolation method used to create the output dataset
flag: -final %s
fine_blur: (a float)
Set the blurring radius to use in the fine resolution
pass to 'x' mm. A small amount (1-2 mm?) of blurring at
the fine step may help with convergence, if there is
some problem, especially if the base volume is very noisy.
[Default == 0 mm = no blurring at the final alignment pass]
flag: -fineblur %f
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
in_matrix: (a file name)
matrix to align input file
flag: -1Dmatrix_apply %s, position: -3
in_param_file: (an existing file name)
Read warp parameters from file and apply them to
the source dataset, and produce a new dataset
flag: -1Dparam_apply %s
interpolation: ('nearestneighbour' or 'linear' or 'cubic' or
'quintic')
Defines interpolation method to use during matching
flag: -interp %s
master: (an existing file name)
Write the output dataset on the same grid as this file
flag: -master %s
newgrid: (a float)
Write the output dataset using isotropic grid spacing in mm
flag: -newgrid %f
nmatch: (an integer (int or long))
Use at most n scattered points to match the datasets.
flag: -nmatch %d
no_pad: (a boolean)
Do not use zero-padding on the base image.
flag: -nopad
nomask: (a boolean)
Don't compute the autoweight/mask; if -weight is not
also used, then every voxel will be counted equally.
flag: -nomask
nwarp: ('bilinear' or 'cubic' or 'quintic' or 'heptic' or 'nonic' or
'poly3' or 'poly5' or 'poly7' or 'poly9')
Experimental nonlinear warping: bilinear or legendre poly.
flag: -nwarp %s
nwarp_fixdep: (a list of items which are 'X' or 'Y' or 'Z' or 'I' or
'J' or 'K')
To fix non-linear warp dependency along directions.
flag: -nwarp_fixdep%s
nwarp_fixmot: (a list of items which are 'X' or 'Y' or 'Z' or 'I' or
'J' or 'K')
To fix motion along directions.
flag: -nwarp_fixmot%s
one_pass: (a boolean)
Use only the refining pass -- do not try a coarse
resolution pass first. Useful if you know that only
small amounts of image alignment are needed.
flag: -onepass
out_file: (a file name)
output file from 3dAllineate
flag: -prefix %s, position: -2
out_matrix: (a file name)
Save the transformation matrix for each volume.
flag: -1Dmatrix_save %s
out_param_file: (a file name)
Save the warp parameters in ASCII (.1D) format.
flag: -1Dparam_save %s
out_weight_file: (a file name)
Write the weight volume to disk as a dataset
flag: -wtprefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
reference: (an existing file name)
file to be used as reference, the first volume will be used
if not given the reference will be the first volume of in_file.
flag: -base %s
replacebase: (a boolean)
If the source has more than one volume, then after the first
volume is aligned to the base
flag: -replacebase
replacemeth: ('leastsq' or 'ls' or 'mutualinfo' or 'mi' or
'corratio_mul' or 'crM' or 'norm_mutualinfo' or 'nmi' or
'hellinger' or 'hel' or 'corratio_add' or 'crA' or 'corratio_uns'
or 'crU')
After first volume is aligned, switch method for later volumes.
For use with '-replacebase'.
flag: -replacemeth %s
source_automask: (an integer (int or long))
Automatically mask the source dataset with dilation or 0.
flag: -source_automask+%d
source_mask: (an existing file name)
mask the input dataset
flag: -source_mask %s
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
two_best: (an integer (int or long))
In the coarse pass, use the best 'bb' set of initial
points to search for the starting point for the fine
pass. If bb==0, then no search is made for the best
starting point, and the identity transformation is
used as the starting point. [Default=5; min=0 max=11]
flag: -twobest %d
two_blur: (a float)
Set the blurring radius for the first pass in mm.
flag: -twoblur
two_first: (a boolean)
Use -twopass on the first image to be registered, and
then on all subsequent images from the source dataset,
use results from the first image's coarse pass to start
the fine pass.
flag: -twofirst
two_pass: (a boolean)
Use a two pass alignment strategy for all volumes, searching
for a large rotation+shift and then refining the alignment.
flag: -twopass
usetemp: (a boolean)
temporary file use
flag: -usetemp
warp_type: ('shift_only' or 'shift_rotate' or 'shift_rotate_scale' or
'affine_general')
Set the warp type.
flag: -warp %s
warpfreeze: (a boolean)
Freeze the non-rigid body parameters after first volume.
flag: -warpfreeze
weight_file: (an existing file name)
Set the weighting for each voxel in the base dataset;
larger weights mean that voxel count more in the cost function.
Must be defined on the same grid as the base dataset
flag: -weight %s
zclip: (a boolean)
Replace negative values in the input datasets (source & base) with
zero.
flag: -zclip
Outputs:
matrix: (a file name)
matrix to align input file
out_file: (a file name)
output image file name
AutoTcorrelate¶
Wraps command 3dAutoTcorrelate
Computes the correlation coefficient between the time series of each pair of voxels in the input dataset, and stores the output into a new anatomical bucket dataset [scaled to shorts to save memory space].
Examples¶
>>> from nipype.interfaces import afni as afni
>>> corr = afni.AutoTcorrelate()
>>> corr.inputs.in_file = 'functional.nii'
>>> corr.inputs.polort = -1
>>> corr.inputs.eta2 = True
>>> corr.inputs.mask = 'mask.nii'
>>> corr.inputs.mask_only_targets = True
>>> corr.cmdline
'3dAutoTcorrelate -eta2 -mask mask.nii -mask_only_targets -prefix functional_similarity_matrix.1D -polort -1 functional.nii'
>>> res = corr.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
timeseries x space (volume or surface) file
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
eta2: (a boolean)
eta^2 similarity
flag: -eta2
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
mask of voxels
flag: -mask %s
mask_only_targets: (a boolean)
use mask only on targets voxels
flag: -mask_only_targets
mutually_exclusive: mask_source
mask_source: (an existing file name)
mask for source voxels
flag: -mask_source %s
mutually_exclusive: mask_only_targets
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
polort: (an integer (int or long))
Remove polynomical trend of order m or -1 for no detrending
flag: -polort %d
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
Autobox¶
Wraps command 3dAutobox
Computes size of a box that fits around the volume. Also can be used to crop the volume to that box.
For complete details, see the `3dAutobox Documentation. <http://afni.nimh.nih.gov/pub/dist/doc/program_help/3dAutobox.html>
Examples¶
>>> from nipype.interfaces import afni as afni
>>> abox = afni.Autobox()
>>> abox.inputs.in_file = 'structural.nii'
>>> abox.inputs.padding = 5
>>> res = abox.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file
flag: -input %s
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
no_clustering: (a boolean)
Don't do any clustering to find box. Any non-zero
voxel will be preserved in the cropped volume.
The default method uses some clustering to find the
cropping box, and will clip off small isolated blobs.
flag: -noclust
out_file: (a file name)
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
padding: (an integer (int or long))
Number of extra voxels to pad on each side of box
flag: -npad %d
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (a file name)
output file
x_max: (an integer (int or long))
x_min: (an integer (int or long))
y_max: (an integer (int or long))
y_min: (an integer (int or long))
z_max: (an integer (int or long))
z_min: (an integer (int or long))
Automask¶
Wraps command 3dAutomask
Create a brain-only mask of the image using AFNI 3dAutomask command
For complete details, see the 3dAutomask Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> automask = afni.Automask()
>>> automask.inputs.in_file = 'functional.nii'
>>> automask.inputs.dilate = 1
>>> automask.inputs.outputtype = "NIFTI"
>>> automask.cmdline
'3dAutomask -apply_prefix functional_masked.nii -dilate 1 -prefix functional_mask.nii functional.nii'
>>> res = automask.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dAutomask
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
brain_file: (a file name)
output file from 3dAutomask
flag: -apply_prefix %s
clfrac: (a float)
sets the clip level fraction (must be 0.1-0.9). A small value will
tend to make the mask larger [default = 0.5].
flag: -clfrac %s
dilate: (an integer (int or long))
dilate the mask outwards
flag: -dilate %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
erode: (an integer (int or long))
erode the mask inwards
flag: -erode %s
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
brain_file: (an existing file name)
brain file (skull stripped)
out_file: (an existing file name)
mask file
Bandpass¶
Wraps command 3dBandpass
Program to lowpass and/or highpass each voxel time series in a dataset, offering more/different options than Fourier
For complete details, see the 3dBandpass Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> from nipype.testing import example_data
>>> bandpass = afni.Bandpass()
>>> bandpass.inputs.in_file = example_data('functional.nii')
>>> bandpass.inputs.highpass = 0.005
>>> bandpass.inputs.lowpass = 0.1
>>> res = bandpass.run()
Inputs:
[Mandatory]
highpass: (a float)
highpass
flag: %f, position: -3
in_file: (an existing file name)
input file to 3dBandpass
flag: %s, position: -1
lowpass: (a float)
lowpass
flag: %f, position: -2
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
automask: (a boolean)
Create a mask from the input dataset
flag: -automask
blur: (a float)
Blur (inside the mask only) with a filter
width (FWHM) of 'fff' millimeters.
flag: -blur %f
despike: (a boolean)
Despike each time series before other processing.
++ Hopefully, you don't actually need to do this,
which is why it is optional.
flag: -despike
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
localPV: (a float)
Replace each vector by the local Principal Vector
(AKA first singular vector) from a neighborhood
of radius 'rrr' millimiters.
++ Note that the PV time series is L2 normalized.
++ This option is mostly for Bob Cox to have fun with.
flag: -localPV %f
mask: (an existing file name)
mask file
flag: -mask %s, position: 2
nfft: (an integer (int or long))
set the FFT length [must be a legal value]
flag: -nfft %d
no_detrend: (a boolean)
Skip the quadratic detrending of the input that
occurs before the FFT-based bandpassing.
++ You would only want to do this if the dataset
had been detrended already in some other program.
flag: -nodetrend
normalize: (a boolean)
Make all output time series have L2 norm = 1
++ i.e., sum of squares = 1
flag: -norm
notrans: (a boolean)
Don't check for initial positive transients in the data:
++ The test is a little slow, so skipping it is OK,
if you KNOW the data time series are transient-free.
flag: -notrans
orthogonalize_dset: (an existing file name)
Orthogonalize each voxel to the corresponding
voxel time series in dataset 'fset', which must
have the same spatial and temporal grid structure
as the main input dataset.
++ At present, only one '-dsort' option is allowed.
flag: -dsort %s
orthogonalize_file: (a list of items which are an existing file name)
Also orthogonalize input to columns in f.1D
++ Multiple '-ort' options are allowed.
flag: -ort %s
out_file: (a file name)
output file from 3dBandpass
flag: -prefix %s, position: 1
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
tr: (a float)
set time step (TR) in sec [default=from dataset header]
flag: -dt %f
Outputs:
out_file: (an existing file name)
output file
BlurInMask¶
Wraps command 3dBlurInMask
Blurs a dataset spatially inside a mask. That’s all. Experimental.
For complete details, see the `3dBlurInMask Documentation. <http://afni.nimh.nih.gov/pub/dist/doc/program_help/3dBlurInMask.html>
Examples¶
>>> from nipype.interfaces import afni as afni
>>> bim = afni.BlurInMask()
>>> bim.inputs.in_file = 'functional.nii'
>>> bim.inputs.mask = 'mask.nii'
>>> bim.inputs.fwhm = 5.0
>>> bim.cmdline
'3dBlurInMask -input functional.nii -FWHM 5.000000 -mask mask.nii -prefix functional_blur'
>>> res = bim.run()
Inputs:
[Mandatory]
fwhm: (a float)
fwhm kernel size
flag: -FWHM %f
in_file: (an existing file name)
input file to 3dSkullStrip
flag: -input %s, position: 1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
automask: (a boolean)
Create an automask from the input dataset.
flag: -automask
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
float_out: (a boolean)
Save dataset as floats, no matter what the input data type is.
flag: -float
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (a file name)
Mask dataset, if desired. Blurring will occur only within the mask.
Voxels NOT in the mask will be set to zero in the output.
flag: -mask %s
multimask: (a file name)
Multi-mask dataset -- each distinct nonzero value in dataset will be
treated as a separate mask for blurring purposes.
flag: -Mmask %s
options: (a string)
options
flag: %s, position: 2
out_file: (a file name)
output to the file
flag: -prefix %s, position: -1
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
preserve: (a boolean)
Normally, voxels not in the mask will be set to zero in the output.
If you want the original values in the dataset to be preserved in
the output, use this option.
flag: -preserve
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
BlurToFWHM¶
Wraps command 3dBlurToFWHM
Blurs a ‘master’ dataset until it reaches a specified FWHM smoothness (approximately).
For complete details, see the to3d Documentation
Examples¶
>>> from nipype.interfaces import afni
>>> blur = afni.preprocess.BlurToFWHM()
>>> blur.inputs.in_file = 'epi.nii'
>>> blur.inputs.fwhm = 2.5
>>> blur.cmdline
'3dBlurToFWHM -FWHM 2.500000 -input epi.nii -prefix epi_afni'
Inputs:
[Mandatory]
in_file: (an existing file name)
The dataset that will be smoothed
flag: -input %s
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
automask: (a boolean)
Create an automask from the input dataset.
flag: -automask
blurmaster: (an existing file name)
The dataset whose smoothness controls the process.
flag: -blurmaster %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
fwhm: (a float)
Blur until the 3D FWHM reaches this value (in mm)
flag: -FWHM %f
fwhmxy: (a float)
Blur until the 2D (x,y)-plane FWHM reaches this value (in mm)
flag: -FWHMxy %f
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
Mask dataset, if desired. Voxels NOT in mask will be set to zero in
output.
flag: -blurmaster %s
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
BrickStat¶
Wraps command 3dBrickStat
Compute maximum and/or minimum voxel values of an input dataset
For complete details, see the 3dBrickStat Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> brickstat = afni.BrickStat()
>>> brickstat.inputs.in_file = 'functional.nii'
>>> brickstat.inputs.mask = 'skeleton_mask.nii.gz'
>>> brickstat.inputs.min = True
>>> res = brickstat.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dmaskave
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
-mask dset = use dset as mask to include/exclude voxels
flag: -mask %s, position: 2
min: (a boolean)
print the minimum value in dataset
flag: -min, position: 1
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
min_val: (a float)
output
Calc¶
Wraps command 3dcalc
This program does voxel-by-voxel arithmetic on 3D datasets
For complete details, see the 3dcalc Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> calc = afni.Calc()
>>> calc.inputs.in_file_a = 'functional.nii'
>>> calc.inputs.in_file_b = 'functional2.nii'
>>> calc.inputs.expr='a*b'
>>> calc.inputs.out_file = 'functional_calc.nii.gz'
>>> calc.inputs.outputtype = "NIFTI"
>>> calc.cmdline
'3dcalc -a functional.nii -b functional2.nii -expr "a*b" -prefix functional_calc.nii.gz'
Inputs:
[Mandatory]
expr: (a string)
expr
flag: -expr "%s", position: 3
in_file_a: (an existing file name)
input file to 3dcalc
flag: -a %s, position: 0
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
in_file_b: (an existing file name)
operand file to 3dcalc
flag: -b %s, position: 1
in_file_c: (an existing file name)
operand file to 3dcalc
flag: -c %s, position: 2
other: (a file name)
other options
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
single_idx: (an integer (int or long))
volume index for in_file_a
start_idx: (an integer (int or long))
start index for in_file_a
requires: stop_idx
stop_idx: (an integer (int or long))
stop index for in_file_a
requires: start_idx
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
ClipLevel¶
Wraps command 3dClipLevel
- Estimates the value at which to clip the anatomical dataset so
- that background regions are set to zero.
For complete details, see the 3dClipLevel Documentation.
Examples¶
>>> from nipype.interfaces.afni import preprocess
>>> cliplevel = preprocess.ClipLevel()
>>> cliplevel.inputs.in_file = 'anatomical.nii'
>>> res = cliplevel.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dClipLevel
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
doall: (a boolean)
Apply the algorithm to each sub-brick separately
flag: -doall, position: 3
mutually_exclusive: g, r, a, d
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
grad: (a file name)
also compute a 'gradual' clip level as a function of voxel position,
and output that to a dataset
flag: -grad %s, position: 3
mutually_exclusive: d, o, a, l, l
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mfrac: (a float)
Use the number ff instead of 0.50 in the algorithm
flag: -mfrac %s, position: 2
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
clip_val: (a float)
output
Copy¶
Wraps command 3dcopy
Copies an image of one type to an image of the same or different type using 3dcopy command
For complete details, see the 3dcopy Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> copy3d = afni.Copy()
>>> copy3d.inputs.in_file = 'functional.nii'
>>> copy3d.cmdline
'3dcopy functional.nii functional_copy'
>>> from copy import deepcopy
>>> copy3d_2 = deepcopy(copy3d)
>>> copy3d_2.inputs.outputtype = 'NIFTI'
>>> copy3d_2.cmdline
'3dcopy functional.nii functional_copy.nii'
>>> copy3d_3 = deepcopy(copy3d)
>>> copy3d_3.inputs.outputtype = 'NIFTI_GZ'
>>> copy3d_3.cmdline
'3dcopy functional.nii functional_copy.nii.gz'
>>> copy3d_4 = deepcopy(copy3d)
>>> copy3d_4.inputs.out_file = 'new_func.nii'
>>> copy3d_4.cmdline
'3dcopy functional.nii new_func.nii'
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dcopy
flag: %s, position: -2
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: %s, position: -1
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
DegreeCentrality¶
Wraps command 3dDegreeCentrality
Performs degree centrality on a dataset using a given maskfile via 3dDegreeCentrality
For complete details, see the `3dDegreeCentrality Documentation. <http://afni.nimh.nih.gov/pub/dist/doc/program_help/3dDegreeCentrality.html>
Examples¶
>>> from nipype.interfaces import afni as afni
>>> degree = afni.DegreeCentrality()
>>> degree.inputs.in_file = 'functional.nii'
>>> degree.inputs.mask = 'mask.nii'
>>> degree.inputs.sparsity = 1 # keep the top one percent of connections
>>> degree.inputs.out_file = 'out.nii'
>>> degree.cmdline
'3dDegreeCentrality -mask mask.nii -prefix out.nii -sparsity 1.000000 functional.nii'
>>> res = degree.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dDegreeCentrality
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
autoclip: (a boolean)
Clip off low-intensity regions in the dataset
flag: -autoclip
automask: (a boolean)
Mask the dataset to target brain-only voxels
flag: -automask
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
mask file to mask input data
flag: -mask %s
oned_file: (a string)
output filepath to text dump of correlation matrix
flag: -out1D %s
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
polort: (an integer (int or long))
flag: -polort %d
sparsity: (a float)
only take the top percent of connections
flag: -sparsity %f
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
thresh: (a float)
threshold to exclude connections where corr <= thresh
flag: -thresh %f
Outputs:
oned_file: (a file name)
The text output of the similarity matrix computedafter thresholding
with one-dimensional and ijk voxel indices, correlations, image
extents, and affine matrix
out_file: (an existing file name)
output file
Despike¶
Wraps command 3dDespike
Removes ‘spikes’ from the 3D+time input dataset
For complete details, see the 3dDespike Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> despike = afni.Despike()
>>> despike.inputs.in_file = 'functional.nii'
>>> despike.cmdline
'3dDespike -prefix functional_despike functional.nii'
>>> res = despike.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dDespike
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
Detrend¶
Wraps command 3dDetrend
This program removes components from voxel time series using linear least squares
For complete details, see the 3dDetrend Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> detrend = afni.Detrend()
>>> detrend.inputs.in_file = 'functional.nii'
>>> detrend.inputs.args = '-polort 2'
>>> detrend.inputs.outputtype = "AFNI"
>>> detrend.cmdline
'3dDetrend -polort 2 -prefix functional_detrend functional.nii'
>>> res = detrend.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dDetrend
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
ECM¶
Wraps command 3dECM
Performs degree centrality on a dataset using a given maskfile via the 3dLFCD command
For complete details, see the `3dECM Documentation. <http://afni.nimh.nih.gov/pub/dist/doc/program_help/3dECM.html>
Examples¶
>>> from nipype.interfaces import afni as afni
>>> ecm = afni.ECM()
>>> ecm.inputs.in_file = 'functional.nii'
>>> ecm.inputs.mask = 'mask.nii'
>>> ecm.inputs.sparsity = 0.1 # keep top 0.1% of connections
>>> ecm.inputs.out_file = 'out.nii'
>>> ecm.cmdline
'3dECM -mask mask.nii -prefix out.nii -sparsity 0.100000 functional.nii'
>>> res = ecm.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dECM
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
autoclip: (a boolean)
Clip off low-intensity regions in the dataset
flag: -autoclip
automask: (a boolean)
Mask the dataset to target brain-only voxels
flag: -automask
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
eps: (a float)
sets the stopping criterion for the power iteration; l2|v_old -
v_new| < eps*|v_old|; default = 0.001
flag: -eps %f
fecm: (a boolean)
Fast centrality method; substantial speed increase but cannot
accomodate thresholding; automatically selected if -thresh or
-sparsity are not set
flag: -fecm
full: (a boolean)
Full power method; enables thresholding; automatically selected if
-thresh or -sparsity are set
flag: -full
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
mask file to mask input data
flag: -mask %s
max_iter: (an integer (int or long))
sets the maximum number of iterations to use in the power iteration;
default = 1000
flag: -max_iter %d
memory: (a float)
Limit memory consumption on system by setting the amount of GB to
limit the algorithm to; default = 2GB
flag: -memory %f
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
polort: (an integer (int or long))
flag: -polort %d
scale: (a float)
scale correlation coefficients in similarity matrix to after
shifting, x >= 0.0; default = 1.0 for -full, 0.5 for -fecm
flag: -scale %f
shift: (a float)
shift correlation coefficients in similarity matrix to enforce non-
negativity, s >= 0.0; default = 0.0 for -full, 1.0 for -fecm
flag: -shift %f
sparsity: (a float)
only take the top percent of connections
flag: -sparsity %f
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
thresh: (a float)
threshold to exclude connections where corr <= thresh
flag: -thresh %f
Outputs:
out_file: (an existing file name)
output file
Eval¶
Wraps command 1deval
Evaluates an expression that may include columns of data from one or more text files
see AFNI Documentation: <http://afni.nimh.nih.gov/pub/dist/doc/program_help/1deval.html>
Examples¶
>>> from nipype.interfaces import afni as afni
>>> eval = afni.Eval()
>>> eval.inputs.in_file_a = 'seed.1D'
>>> eval.inputs.in_file_b = 'resp.1D'
>>> eval.inputs.expr='a*b'
>>> eval.inputs.out1D = True
>>> eval.inputs.out_file = 'data_calc.1D'
>>> calc.cmdline
'3deval -a timeseries1.1D -b timeseries2.1D -expr "a*b" -1D -prefix data_calc.1D'
Inputs:
[Mandatory]
expr: (a string)
expr
flag: -expr "%s", position: 3
in_file_a: (an existing file name)
input file to 1deval
flag: -a %s, position: 0
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
in_file_b: (an existing file name)
operand file to 1deval
flag: -b %s, position: 1
in_file_c: (an existing file name)
operand file to 1deval
flag: -c %s, position: 2
other: (a file name)
other options
out1D: (a boolean)
output in 1D
flag: -1D
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
single_idx: (an integer (int or long))
volume index for in_file_a
start_idx: (an integer (int or long))
start index for in_file_a
requires: stop_idx
stop_idx: (an integer (int or long))
stop index for in_file_a
requires: start_idx
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
FWHMx¶
Wraps command 3dFWHMx
Unlike the older 3dFWHM, this program computes FWHMs for all sub-bricks in the input dataset, each one separately. The output for each one is written to the file specified by ‘-out’. The mean (arithmetic or geometric) of all the FWHMs along each axis is written to stdout. (A non-positive output value indicates something bad happened; e.g., FWHM in z is meaningless for a 2D dataset; the estimation method computed incoherent intermediate results.)
Examples¶
>>> from nipype.interfaces import afni as afp
>>> fwhm = afp.FWHMx()
>>> fwhm.inputs.in_file = 'functional.nii'
>>> fwhm.cmdline
'3dFWHMx -input functional.nii -out functional_subbricks.out > functional_fwhmx.out'
(Classic) METHOD:
- Calculate ratio of variance of first differences to data variance.
- Should be the same as 3dFWHM for a 1-brick dataset. (But the output format is simpler to use in a script.)
Note
IMPORTANT NOTE [AFNI > 16]
A completely new method for estimating and using noise smoothness values is now available in 3dFWHMx and 3dClustSim. This method is implemented in the ‘-acf’ options to both programs. ‘ACF’ stands for (spatial) AutoCorrelation Function, and it is estimated by calculating moments of differences out to a larger radius than before.
Notably, real FMRI data does not actually have a Gaussian-shaped ACF, so the estimated ACF is then fit (in 3dFWHMx) to a mixed model (Gaussian plus mono-exponential) of the form
where  is the radius, and
is the radius, and 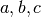 are the fitted parameters.
The apparent FWHM from this model is usually somewhat larger in real data
than the FWHM estimated from just the nearest-neighbor differences used
in the ‘classic’ analysis.
are the fitted parameters.
The apparent FWHM from this model is usually somewhat larger in real data
than the FWHM estimated from just the nearest-neighbor differences used
in the ‘classic’ analysis.
The longer tails provided by the mono-exponential are also significant. 3dClustSim has also been modified to use the ACF model given above to generate noise random fields.
Note
TL;DR or summary
The take-awaymessage is that the ‘classic’ 3dFWHMx and 3dClustSim analysis, using a pure Gaussian ACF, is not very correct for FMRI data – I cannot speak for PET or MEG data.
Warning
Do NOT use 3dFWHMx on the statistical results (e.g., ‘-bucket’) from 3dDeconvolve or 3dREMLfit!!! The function of 3dFWHMx is to estimate the smoothness of the time series NOISE, not of the statistics. This proscription is especially true if you plan to use 3dClustSim next!!
Note
Recommendations
- For FMRI statistical purposes, you DO NOT want the FWHM to reflect the spatial structure of the underlying anatomy. Rather, you want the FWHM to reflect the spatial structure of the noise. This means that the input dataset should not have anatomical (spatial) structure.
- One good form of input is the output of ‘3dDeconvolve -errts’, which is the dataset of residuals left over after the GLM fitted signal model is subtracted out from each voxel’s time series.
- If you don’t want to go to that much trouble, use ‘-detrend’ to approximately subtract out the anatomical spatial structure, OR use the output of 3dDetrend for the same purpose.
- If you do not use ‘-detrend’, the program attempts to find non-zero spatial structure in the input, and will print a warning message if it is detected.
Note
Notes on -demend
- I recommend this option, and it is not the default only for historical compatibility reasons. It may become the default someday.
- It is already the default in program 3dBlurToFWHM. This is the same detrending as done in 3dDespike; using 2*q+3 basis functions for q > 0.
- If you don’t use ‘-detrend’, the program now [Aug 2010] checks if a large number of voxels are have significant nonzero means. If so, the program will print a warning message suggesting the use of ‘-detrend’, since inherent spatial structure in the image will bias the estimation of the FWHM of the image time series NOISE (which is usually the point of using 3dFWHMx).
Inputs:
[Mandatory]
in_file: (an existing file name)
input dataset
flag: -input %s
[Optional]
acf: (a boolean or a file name or a tuple of the form: (an existing
file name, a float), nipype default value: False)
computes the spatial autocorrelation
flag: -acf
args: (a string)
Additional parameters to the command
flag: %s
arith: (a boolean)
if in_file has more than one sub-brick, compute the final estimate
asthe arithmetic mean of the individual sub-brick FWHM estimates
flag: -arith
mutually_exclusive: geom
automask: (a boolean, nipype default value: False)
compute a mask from THIS dataset, a la 3dAutomask
flag: -automask
combine: (a boolean)
combine the final measurements along each axis
flag: -combine
compat: (a boolean)
be compatible with the older 3dFWHM
flag: -compat
demed: (a boolean)
If the input dataset has more than one sub-brick (e.g., has a time
axis), then subtract the median of each voxel's time series before
processing FWHM. This will tend to remove intrinsic spatial
structure and leave behind the noise.
flag: -demed
mutually_exclusive: detrend
detrend: (a boolean or an integer (int or long), nipype default
value: False)
instead of demed (0th order detrending), detrend to the specified
order. If order is not given, the program picks q=NT/30. -detrend
disables -demed, and includes -unif.
flag: -detrend
mutually_exclusive: demed
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
geom: (a boolean)
if in_file has more than one sub-brick, compute the final estimate
asthe geometric mean of the individual sub-brick FWHM estimates
flag: -geom
mutually_exclusive: arith
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
use only voxels that are nonzero in mask
flag: -mask %s
out_detrend: (a file name)
Save the detrended file into a dataset
flag: -detprefix %s
out_file: (a file name)
output file
flag: > %s, position: -1
out_subbricks: (a file name)
output file listing the subbricks FWHM
flag: -out %s
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
unif: (a boolean)
If the input dataset has more than one sub-brick, then normalize
each voxel's time series to have the same MAD before processing
FWHM.
flag: -unif
Outputs:
acf_param: (a tuple of the form: (a float, a float, a float) or a
tuple of the form: (a float, a float, a float, a float))
fitted ACF model parameters
fwhm: (a tuple of the form: (a float, a float, a float) or a tuple of
the form: (a float, a float, a float, a float))
FWHM along each axis
out_acf: (an existing file name)
output acf file
out_detrend: (a file name)
output file, detrended
out_file: (an existing file name)
output file
out_subbricks: (an existing file name)
output file (subbricks)
Fim¶
Wraps command 3dfim+
Program to calculate the cross-correlation of an ideal reference waveform with the measured FMRI time series for each voxel
For complete details, see the 3dfim+ Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> fim = afni.Fim()
>>> fim.inputs.in_file = 'functional.nii'
>>> fim.inputs.ideal_file= 'seed.1D'
>>> fim.inputs.out_file = 'functional_corr.nii'
>>> fim.inputs.out = 'Correlation'
>>> fim.inputs.fim_thr = 0.0009
>>> res = fim.run()
Inputs:
[Mandatory]
ideal_file: (an existing file name)
ideal time series file name
flag: -ideal_file %s, position: 2
in_file: (an existing file name)
input file to 3dfim+
flag: -input %s, position: 1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
fim_thr: (a float)
fim internal mask threshold value
flag: -fim_thr %f, position: 3
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out: (a string)
Flag to output the specified parameter
flag: -out %s, position: 4
out_file: (a file name)
output image file name
flag: -bucket %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
Fourier¶
Wraps command 3dFourier
Program to lowpass and/or highpass each voxel time series in a dataset, via the FFT
For complete details, see the 3dFourier Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> fourier = afni.Fourier()
>>> fourier.inputs.in_file = 'functional.nii'
>>> fourier.inputs.args = '-retrend'
>>> fourier.inputs.highpass = 0.005
>>> fourier.inputs.lowpass = 0.1
>>> res = fourier.run()
Inputs:
[Mandatory]
highpass: (a float)
highpass
flag: -highpass %f, position: 1
in_file: (an existing file name)
input file to 3dFourier
flag: %s, position: -1
lowpass: (a float)
lowpass
flag: -lowpass %f, position: 0
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
Hist¶
Wraps command 3dHist
Computes average of all voxels in the input dataset which satisfy the criterion in the options list
For complete details, see the 3dHist Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> hist = afni.Hist()
>>> hist.inputs.in_file = 'functional.nii'
>>> hist.cmdline
'3dHist -input functional.nii -prefix functional_hist'
>>> res = hist.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dHist
flag: -input %s, position: 1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
bin_width: (a float)
bin width
flag: -binwidth %f
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
matrix to align input file
flag: -mask %s
max_value: (a float)
maximum intensity value
flag: -max %f
min_value: (a float)
minimum intensity value
flag: -min %f
nbin: (an integer (int or long))
number of bins
flag: -nbin %d
out_file: (a file name)
Write histogram to niml file with this prefix
flag: -prefix %s
out_show: (a file name)
output image file name
flag: > %s, position: -1
showhist: (a boolean, nipype default value: False)
write a text visual histogram
flag: -showhist
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
out_show: (a file name)
output visual histogram
LFCD¶
Wraps command 3dLFCD
Performs degree centrality on a dataset using a given maskfile via the 3dLFCD command
For complete details, see the `3dLFCD Documentation. <http://afni.nimh.nih.gov/pub/dist/doc/program_help/3dLFCD.html>
Examples¶
>>> from nipype.interfaces import afni as afni
>>> lfcd = afni.LFCD()
>>> lfcd.inputs.in_file = 'functional.nii'
>>> lfcd.inputs.mask = 'mask.nii'
>>> lfcd.inputs.thresh = 0.8 # keep all connections with corr >= 0.8
>>> lfcd.inputs.out_file = 'out.nii'
>>> lfcd.cmdline
'3dLFCD -mask mask.nii -prefix out.nii -thresh 0.800000 functional.nii'
>>> res = lfcd.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dLFCD
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
autoclip: (a boolean)
Clip off low-intensity regions in the dataset
flag: -autoclip
automask: (a boolean)
Mask the dataset to target brain-only voxels
flag: -automask
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
mask file to mask input data
flag: -mask %s
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
polort: (an integer (int or long))
flag: -polort %d
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
thresh: (a float)
threshold to exclude connections where corr <= thresh
flag: -thresh %f
Outputs:
out_file: (an existing file name)
output file
MaskTool¶
Wraps command 3dmask_tool
3dmask_tool - for combining/dilating/eroding/filling masks
For complete details, see the 3dmask_tool Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> automask = afni.Automask()
>>> automask.inputs.in_file = 'functional.nii'
>>> automask.inputs.dilate = 1
>>> automask.inputs.outputtype = "NIFTI"
>>> automask.cmdline
'3dAutomask -apply_prefix functional_masked.nii -dilate 1 -prefix functional_mask.nii functional.nii'
>>> res = automask.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file or files to 3dmask_tool
flag: -input %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
count: (a boolean)
Instead of created a binary 0/1 mask dataset, create one with.
counts of voxel overlap, i.e each voxel will contain the number of
masks that it is set in.
flag: -count, position: 2
datum: ('byte' or 'short' or 'float')
specify data type for output. Valid types are 'byte', 'short' and
'float'.
flag: -datum %s
dilate_inputs: (a string)
Use this option to dilate and/or erode datasets as they are read.
ex. '5 -5' to dilate and erode 5 times
flag: -dilate_inputs %s
dilate_results: (a string)
dilate and/or erode combined mask at the given levels.
flag: -dilate_results %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
fill_dirs: (a string)
fill holes only in the given directions. This option is for use with
-fill holes. should be a single string that specifies 1-3 of the
axes using {x,y,z} labels (i.e. dataset axis order), or using the
labels in {R,L,A,P,I,S}.
flag: -fill_dirs %s
requires: fill_holes
fill_holes: (a boolean)
This option can be used to fill holes in the resulting mask, i.e.
after all other processing has been done.
flag: -fill_holes
frac: (a float)
When combining masks (across datasets and sub-bricks), use this
option to restrict the result to a certain fraction of the set of
volumes
flag: -frac %s
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
inter: (a boolean)
intersection, this means -frac 1.0
flag: -inter
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
union: (a boolean)
union, this means -frac 0
flag: -union
Outputs:
out_file: (an existing file name)
mask file
Maskave¶
Wraps command 3dmaskave
Computes average of all voxels in the input dataset which satisfy the criterion in the options list
For complete details, see the 3dmaskave Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> maskave = afni.Maskave()
>>> maskave.inputs.in_file = 'functional.nii'
>>> maskave.inputs.mask= 'seed_mask.nii'
>>> maskave.inputs.quiet= True
>>> maskave.cmdline
'3dmaskave -mask seed_mask.nii -quiet functional.nii > functional_maskave.1D'
>>> res = maskave.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dmaskave
flag: %s, position: -2
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
matrix to align input file
flag: -mask %s, position: 1
out_file: (a file name)
output image file name
flag: > %s, position: -1
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
quiet: (a boolean)
matrix to align input file
flag: -quiet, position: 2
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
Means¶
Wraps command 3dMean
Takes the voxel-by-voxel mean of all input datasets using 3dMean
see AFNI Documentation: <http://afni.nimh.nih.gov/pub/dist/doc/program_help/3dMean.html>
Examples¶
>>> from nipype.interfaces import afni as afni
>>> means = afni.Means()
>>> means.inputs.in_file_a = 'im1.nii'
>>> means.inputs.in_file_b = 'im2.nii'
>>> means.inputs.out_file = 'output.nii'
>>> means.cmdline
'3dMean im1.nii im2.nii -prefix output.nii'
Inputs:
[Mandatory]
in_file_a: (an existing file name)
input file to 3dMean
flag: %s, position: 0
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
count: (a boolean)
compute count of non-zero voxels
flag: -count
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
in_file_b: (an existing file name)
another input file to 3dMean
flag: %s, position: 1
mask_inter: (a boolean)
create intersection mask
flag: -mask_inter
mask_union: (a boolean)
create union mask
flag: -mask_union
non_zero: (a boolean)
use only non-zero values
flag: -non_zero
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
scale: (a string)
scaling of output
flag: -%sscale
sqr: (a boolean)
mean square instead of value
flag: -sqr
std_dev: (a boolean)
calculate std dev
flag: -stdev
summ: (a boolean)
take sum, (not average)
flag: -sum
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
Merge¶
Wraps command 3dmerge
Merge or edit volumes using AFNI 3dmerge command
For complete details, see the 3dmerge Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> merge = afni.Merge()
>>> merge.inputs.in_files = ['functional.nii', 'functional2.nii']
>>> merge.inputs.blurfwhm = 4
>>> merge.inputs.doall = True
>>> merge.inputs.out_file = 'e7.nii'
>>> res = merge.run()
Inputs:
[Mandatory]
in_files: (a list of items which are an existing file name)
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
blurfwhm: (an integer (int or long))
FWHM blur value (mm)
flag: -1blur_fwhm %d
doall: (a boolean)
apply options to all sub-bricks in dataset
flag: -doall
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
OutlierCount¶
Wraps command 3dToutcount
Create a 3D dataset from 2D image files using AFNI to3d command
For complete details, see the to3d Documentation
Examples¶
>>> from nipype.interfaces import afni
>>> toutcount = afni.OutlierCount()
>>> toutcount.inputs.in_file = 'functional.nii'
>>> toutcount.cmdline
'3dToutcount functional.nii > functional_outliers'
>>> res = toutcount.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input dataset
flag: %s, position: -2
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
autoclip: (a boolean, nipype default value: False)
clip off small voxels
flag: -autoclip
mutually_exclusive: in_file
automask: (a boolean, nipype default value: False)
clip off small voxels
flag: -automask
mutually_exclusive: in_file
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
fraction: (a boolean, nipype default value: False)
write out the fraction of masked voxels which are outliers at each
timepoint
flag: -fraction
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
interval: (a boolean, nipype default value: False)
write out the median + 3.5 MAD of outlier count with each timepoint
flag: -range
legendre: (a boolean, nipype default value: False)
use Legendre polynomials
flag: -legendre
mask: (an existing file name)
only count voxels within the given mask
flag: -mask %s
mutually_exclusive: autoclip, automask
out_file: (a file name)
capture standard output
flag: > %s, position: -1
outliers_file: (a file name)
output image file name
flag: -save %s
polort: (an integer (int or long))
detrend each voxel timeseries with polynomials
flag: -polort %d
qthr: (0.0 <= a floating point number <= 1.0)
indicate a value for q to compute alpha
flag: -qthr %.5f
save_outliers: (a boolean, nipype default value: False)
enables out_file option
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (a file name)
capture standard output
flag: > %s, position: -1
out_outliers: (an existing file name)
output image file name
QualityIndex¶
Wraps command 3dTqual
Create a 3D dataset from 2D image files using AFNI to3d command
For complete details, see the to3d Documentation
Examples¶
>>> from nipype.interfaces import afni
>>> tqual = afni.QualityIndex()
>>> tqual.inputs.in_file = 'functional.nii'
>>> tqual.cmdline
'3dTqual functional.nii > functional_tqual'
>>> res = tqual.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input dataset
flag: %s, position: -2
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
autoclip: (a boolean, nipype default value: False)
clip off small voxels
flag: -autoclip
mutually_exclusive: mask
automask: (a boolean, nipype default value: False)
clip off small voxels
flag: -automask
mutually_exclusive: mask
clip: (a float)
clip off values below
flag: -clip %f
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
interval: (a boolean, nipype default value: False)
write out the median + 3.5 MAD of outlier count with each timepoint
flag: -range
mask: (an existing file name)
compute correlation only across masked voxels
flag: -mask %s
mutually_exclusive: autoclip, automask
out_file: (a file name)
capture standard output
flag: > %s, position: -1
quadrant: (a boolean, nipype default value: False)
Similar to -spearman, but using 1 minus the quadrant correlation
coefficient as the quality index.
flag: -quadrant
spearman: (a boolean, nipype default value: False)
Quality index is 1 minus the Spearman (rank) correlation coefficient
of each sub-brick with the median sub-brick. (default)
flag: -spearman
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (a file name)
file containing the caputured standard output
ROIStats¶
Wraps command 3dROIstats
Display statistics over masked regions
For complete details, see the 3dROIstats Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> roistats = afni.ROIStats()
>>> roistats.inputs.in_file = 'functional.nii'
>>> roistats.inputs.mask = 'skeleton_mask.nii.gz'
>>> roistats.inputs.quiet=True
>>> res = roistats.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dROIstats
flag: %s, position: -1
terminal_output: ('allatonce', nipype default value: allatonce)
Control terminal output:`allatonce` - waits till command is finished
to display output
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
input mask
flag: -mask %s, position: 3
mask_f2short: (a boolean)
Tells the program to convert a float mask to short integers, by
simple rounding.
flag: -mask_f2short, position: 2
quiet: (a boolean)
execute quietly
flag: -quiet, position: 1
Outputs:
stats: (an existing file name)
output tab separated values file
Refit¶
Wraps command 3drefit
Changes some of the information inside a 3D dataset’s header
For complete details, see the `3drefit Documentation. <http://afni.nimh.nih.gov/pub/dist/doc/program_help/3drefit.html>
Examples¶
>>> from nipype.interfaces import afni as afni
>>> refit = afni.Refit()
>>> refit.inputs.in_file = 'structural.nii'
>>> refit.inputs.deoblique = True
>>> refit.cmdline
'3drefit -deoblique structural.nii'
>>> res = refit.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3drefit
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
deoblique: (a boolean)
replace current transformation matrix with cardinal matrix
flag: -deoblique
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
space: ('TLRC' or 'MNI' or 'ORIG')
Associates the dataset with a specific template type, e.g. TLRC,
MNI, ORIG
flag: -space %s
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
xdel: (a float)
new x voxel dimension in mm
flag: -xdel %f
xorigin: (a string)
x distance for edge voxel offset
flag: -xorigin %s
ydel: (a float)
new y voxel dimension in mm
flag: -ydel %f
yorigin: (a string)
y distance for edge voxel offset
flag: -yorigin %s
zdel: (a float)
new z voxel dimension in mm
flag: -zdel %f
zorigin: (a string)
z distance for edge voxel offset
flag: -zorigin %s
Outputs:
out_file: (an existing file name)
output file
Resample¶
Wraps command 3dresample
Resample or reorient an image using AFNI 3dresample command
For complete details, see the 3dresample Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> resample = afni.Resample()
>>> resample.inputs.in_file = 'functional.nii'
>>> resample.inputs.orientation= 'RPI'
>>> resample.inputs.outputtype = "NIFTI"
>>> resample.cmdline
'3dresample -orient RPI -prefix functional_resample.nii -inset functional.nii'
>>> res = resample.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dresample
flag: -inset %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
master: (a file name)
align dataset grid to a reference file
flag: -master %s
orientation: (a string)
new orientation code
flag: -orient %s
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
resample_mode: ('NN' or 'Li' or 'Cu' or 'Bk')
resampling method from set {'NN', 'Li', 'Cu', 'Bk'}. These are for
'Nearest Neighbor', 'Linear', 'Cubic' and 'Blocky' interpolation,
respectively. Default is NN.
flag: -rmode %s
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
voxel_size: (a tuple of the form: (a float, a float, a float))
resample to new dx, dy and dz
flag: -dxyz %f %f %f
Outputs:
out_file: (an existing file name)
output file
Retroicor¶
Wraps command 3dretroicor
Performs Retrospective Image Correction for physiological motion effects, using a slightly modified version of the RETROICOR algorithm
The durations of the physiological inputs are assumed to equal the duration of the dataset. Any constant sampling rate may be used, but 40 Hz seems to be acceptable. This program’s cardiac peak detection algorithm is rather simplistic, so you might try using the scanner’s cardiac gating output (transform it to a spike wave if necessary).
This program uses slice timing information embedded in the dataset to estimate the proper cardiac/respiratory phase for each slice. It makes sense to run this program before any program that may destroy the slice timings (e.g. 3dvolreg for motion correction).
For complete details, see the 3dretroicor Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> ret = afni.Retroicor()
>>> ret.inputs.in_file = 'functional.nii'
>>> ret.inputs.card = 'mask.1D'
>>> ret.inputs.resp = 'resp.1D'
>>> res = ret.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dretroicor
flag: %s, position: -1
out_file: (a file name)
output image file name
flag: -prefix %s, position: 1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
card: (an existing file name)
1D cardiac data file for cardiac correction
flag: -card %s, position: -2
cardphase: (a file name)
Filename for 1D cardiac phase output
flag: -cardphase %s, position: -6
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
order: (an integer (int or long))
The order of the correction (2 is typical)
flag: -order %s, position: -5
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
resp: (an existing file name)
1D respiratory waveform data for correction
flag: -resp %s, position: -3
respphase: (a file name)
Filename for 1D resp phase output
flag: -respphase %s, position: -7
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
threshold: (an integer (int or long))
Threshold for detection of R-wave peaks in input (Make sure it is
above the background noise level, Try 3/4 or 4/5 times range plus
minimum)
flag: -threshold %d, position: -4
Outputs:
out_file: (an existing file name)
output file
Seg¶
Wraps command 3dSeg
- 3dSeg segments brain volumes into tissue classes. The program allows
- for adding a variety of global and voxelwise priors. However for the moment, only mixing fractions and MRF are documented.
For complete details, see the `3dSeg Documentation. <https://afni.nimh.nih.gov/pub/dist/doc/program_help/3dSeg.html>
Examples¶
>>> from nipype.interfaces.afni import preprocess
>>> seg = preprocess.Seg()
>>> seg.inputs.in_file = 'structural.nii'
>>> seg.inputs.mask = 'AUTO'
>>> res = seg.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
ANAT is the volume to segment
flag: -anat %s, position: -1
mask: ('AUTO' or an existing file name)
only non-zero voxels in mask are analyzed. mask can either be a
dataset or the string "AUTO" which would use AFNI's automask
function to create the mask.
flag: -mask %s, position: -2
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
bias_classes: (a string)
A semcolon demlimited string of classes that contribute to the
estimation of the bias field
flag: -bias_classes %s
bias_fwhm: (a float)
The amount of blurring used when estimating the field bias with the
Wells method
flag: -bias_fwhm %f
blur_meth: ('BFT' or 'BIM')
set the blurring method for bias field estimation
flag: -blur_meth %s
bmrf: (a float)
Weighting factor controlling spatial homogeneity of the
classifications
flag: -bmrf %f
classes: (a string)
CLASS_STRING is a semicolon delimited string of class labels
flag: -classes %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
main_N: (an integer (int or long))
Number of iterations to perform.
flag: -main_N %d
mixfloor: (a float)
Set the minimum value for any class's mixing fraction
flag: -mixfloor %f
mixfrac: (a string)
MIXFRAC sets up the volume-wide (within mask) tissue fractions while
initializing the segmentation (see IGNORE for exception)
flag: -mixfrac %s
prefix: (a string)
the prefix for the output folder containing all output volumes
flag: -prefix %s
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
SkullStrip¶
Wraps command 3dSkullStrip
A program to extract the brain from surrounding tissue from MRI T1-weighted images
For complete details, see the 3dSkullStrip Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> skullstrip = afni.SkullStrip()
>>> skullstrip.inputs.in_file = 'functional.nii'
>>> skullstrip.inputs.args = '-o_ply'
>>> res = skullstrip.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dSkullStrip
flag: -input %s, position: 1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
TCat¶
Wraps command 3dTcat
Concatenate sub-bricks from input datasets into one big 3D+time dataset
For complete details, see the 3dTcat Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> tcat = afni.TCat()
>>> tcat.inputs.in_files = ['functional.nii', 'functional2.nii']
>>> tcat.inputs.out_file= 'functional_tcat.nii'
>>> tcat.inputs.rlt = '+'
>>> res = tcat.run()
Inputs:
[Mandatory]
in_files: (a list of items which are an existing file name)
input file to 3dTcat
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
rlt: (a string)
options
flag: -rlt%s, position: 1
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
TCorr1D¶
Wraps command 3dTcorr1D
Computes the correlation coefficient between each voxel time series in the input 3D+time dataset. For complete details, see the 3dTcorr1D Documentation.
>>> from nipype.interfaces import afni as afni
>>> tcorr1D = afni.TCorr1D()
>>> tcorr1D.inputs.xset= 'u_rc1s1_Template.nii'
>>> tcorr1D.inputs.y_1d = 'seed.1D'
>>> tcorr1D.cmdline
'3dTcorr1D -prefix u_rc1s1_Template_correlation.nii.gz u_rc1s1_Template.nii seed.1D'
>>> res = tcorr1D.run()
Inputs:
[Mandatory]
xset: (an existing file name)
3d+time dataset input
flag: %s, position: -2
y_1d: (an existing file name)
1D time series file input
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
ktaub: (a boolean)
Correlation is the Kendall's tau_b correlation coefficient
flag: -ktaub, position: 1
mutually_exclusive: pearson, spearman, quadrant
out_file: (a file name)
output filename prefix
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
pearson: (a boolean)
Correlation is the normal Pearson correlation coefficient
flag: -pearson, position: 1
mutually_exclusive: spearman, quadrant, ktaub
quadrant: (a boolean)
Correlation is the quadrant correlation coefficient
flag: -quadrant, position: 1
mutually_exclusive: pearson, spearman, ktaub
spearman: (a boolean)
Correlation is the Spearman (rank) correlation coefficient
flag: -spearman, position: 1
mutually_exclusive: pearson, quadrant, ktaub
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file containing correlations
TCorrMap¶
Wraps command 3dTcorrMap
For each voxel time series, computes the correlation between it and all other voxels, and combines this set of values into the output dataset(s) in some way.
For complete details, see the `3dTcorrMap Documentation. <http://afni.nimh.nih.gov/pub/dist/doc/program_help/3dTcorrMap.html>
Examples¶
>>> from nipype.interfaces import afni as afni
>>> tcm = afni.TCorrMap()
>>> tcm.inputs.in_file = 'functional.nii'
>>> tcm.inputs.mask = 'mask.nii'
>>> tcm.mean_file = '%s_meancorr.nii'
>>> res = tcm.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
flag: -input %s
[Optional]
absolute_threshold: (a file name)
flag: -Thresh %f %s
mutually_exclusive: absolute_threshold, var_absolute_threshold,
var_absolute_threshold_normalize
args: (a string)
Additional parameters to the command
flag: %s
automask: (a boolean)
flag: -automask
average_expr: (a file name)
flag: -Aexpr %s %s
mutually_exclusive: average_expr, average_expr_nonzero, sum_expr
average_expr_nonzero: (a file name)
flag: -Cexpr %s %s
mutually_exclusive: average_expr, average_expr_nonzero, sum_expr
bandpass: (a tuple of the form: (a float, a float))
flag: -bpass %f %f
blur_fwhm: (a float)
flag: -Gblur %f
correlation_maps: (a file name)
flag: -CorrMap %s
correlation_maps_masked: (a file name)
flag: -CorrMask %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
expr: (a string)
histogram: (a file name)
flag: -Hist %d %s
histogram_bin_numbers: (an integer (int or long))
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
flag: -mask %s
mean_file: (a file name)
flag: -Mean %s
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
pmean: (a file name)
flag: -Pmean %s
polort: (an integer (int or long))
flag: -polort %d
qmean: (a file name)
flag: -Qmean %s
regress_out_timeseries: (a file name)
flag: -ort %s
seeds: (an existing file name)
flag: -seed %s
mutually_exclusive: s, e, e, d, s, _, w, i, d, t, h
seeds_width: (a float)
flag: -Mseed %f
mutually_exclusive: s, e, e, d, s
sum_expr: (a file name)
flag: -Sexpr %s %s
mutually_exclusive: average_expr, average_expr_nonzero, sum_expr
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
thresholds: (a list of items which are an integer (int or long))
var_absolute_threshold: (a file name)
flag: -VarThresh %f %f %f %s
mutually_exclusive: absolute_threshold, var_absolute_threshold,
var_absolute_threshold_normalize
var_absolute_threshold_normalize: (a file name)
flag: -VarThreshN %f %f %f %s
mutually_exclusive: absolute_threshold, var_absolute_threshold,
var_absolute_threshold_normalize
zmean: (a file name)
flag: -Zmean %s
Outputs:
absolute_threshold: (a file name)
average_expr: (a file name)
average_expr_nonzero: (a file name)
correlation_maps: (a file name)
correlation_maps_masked: (a file name)
histogram: (a file name)
mean_file: (a file name)
pmean: (a file name)
qmean: (a file name)
sum_expr: (a file name)
var_absolute_threshold: (a file name)
var_absolute_threshold_normalize: (a file name)
zmean: (a file name)
TCorrelate¶
Wraps command 3dTcorrelate
Computes the correlation coefficient between corresponding voxel time series in two input 3D+time datasets ‘xset’ and ‘yset’
For complete details, see the 3dTcorrelate Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> tcorrelate = afni.TCorrelate()
>>> tcorrelate.inputs.xset= 'u_rc1s1_Template.nii'
>>> tcorrelate.inputs.yset = 'u_rc1s2_Template.nii'
>>> tcorrelate.inputs.out_file = 'functional_tcorrelate.nii.gz'
>>> tcorrelate.inputs.polort = -1
>>> tcorrelate.inputs.pearson = True
>>> res = tcarrelate.run()
Inputs:
[Mandatory]
xset: (an existing file name)
input xset
flag: %s, position: -2
yset: (an existing file name)
input yset
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
pearson: (a boolean)
Correlation is the normal Pearson correlation coefficient
flag: -pearson, position: 1
polort: (an integer (int or long))
Remove polynomical trend of order m
flag: -polort %d, position: 2
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
TShift¶
Wraps command 3dTshift
Shifts voxel time series from input so that seperate slices are aligned to the same temporal origin
For complete details, see the `3dTshift Documentation. <http://afni.nimh.nih.gov/pub/dist/doc/program_help/3dTshift.html>
Examples¶
>>> from nipype.interfaces import afni as afni
>>> tshift = afni.TShift()
>>> tshift.inputs.in_file = 'functional.nii'
>>> tshift.inputs.tpattern = 'alt+z'
>>> tshift.inputs.tzero = 0.0
>>> tshift.cmdline #doctest:
'3dTshift -prefix functional_tshift -tpattern alt+z -tzero 0.0 functional.nii'
>>> res = tshift.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dTShift
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore: (an integer (int or long))
ignore the first set of points specified
flag: -ignore %s
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
interp: ('Fourier' or 'linear' or 'cubic' or 'quintic' or 'heptic')
different interpolation methods (see 3dTShift for details) default =
Fourier
flag: -%s
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
rlt: (a boolean)
Before shifting, remove the mean and linear trend
flag: -rlt
rltplus: (a boolean)
Before shifting, remove the mean and linear trend and later put back
the mean
flag: -rlt+
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
tpattern: (a string)
use specified slice time pattern rather than one in header
flag: -tpattern %s
tr: (a string)
manually set the TRYou can attach suffix "s" for seconds or "ms" for
milliseconds.
flag: -TR %s
tslice: (an integer (int or long))
align each slice to time offset of given slice
flag: -slice %s
mutually_exclusive: tzero
tzero: (a float)
align each slice to given time offset
flag: -tzero %s
mutually_exclusive: tslice
Outputs:
out_file: (an existing file name)
output file
TStat¶
Wraps command 3dTstat
Compute voxel-wise statistics using AFNI 3dTstat command
For complete details, see the 3dTstat Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> tstat = afni.TStat()
>>> tstat.inputs.in_file = 'functional.nii'
>>> tstat.inputs.args= '-mean'
>>> tstat.inputs.out_file = "stats"
>>> tstat.cmdline
'3dTstat -mean -prefix stats functional.nii'
>>> res = tstat.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dTstat
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
mask file
flag: -mask %s
options: (a string)
selected statistical output
flag: %s
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
To3D¶
Wraps command to3d
Create a 3D dataset from 2D image files using AFNI to3d command
For complete details, see the to3d Documentation
Examples¶
>>> from nipype.interfaces import afni
>>> To3D = afni.To3D()
>>> To3D.inputs.datatype = 'float'
>>> To3D.inputs.in_folder = '.'
>>> To3D.inputs.out_file = 'dicomdir.nii'
>>> To3D.inputs.filetype = "anat"
>>> To3D.cmdline
'to3d -datum float -anat -prefix dicomdir.nii ./*.dcm'
>>> res = To3D.run()
Inputs:
[Mandatory]
in_folder: (an existing directory name)
folder with DICOM images to convert
flag: %s/*.dcm, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
assumemosaic: (a boolean)
assume that Siemens image is mosaic
flag: -assume_dicom_mosaic
datatype: ('short' or 'float' or 'byte' or 'complex')
set output file datatype
flag: -datum %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
filetype: ('spgr' or 'fse' or 'epan' or 'anat' or 'ct' or 'spct' or
'pet' or 'mra' or 'bmap' or 'diff' or 'omri' or 'abuc' or 'fim' or
'fith' or 'fico' or 'fitt' or 'fift' or 'fizt' or 'fict' or 'fibt'
or 'fibn' or 'figt' or 'fipt' or 'fbuc')
type of datafile being converted
flag: -%s
funcparams: (a string)
parameters for functional data
flag: -time:zt %s alt+z2
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
skipoutliers: (a boolean)
skip the outliers check
flag: -skip_outliers
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
Volreg¶
Wraps command 3dvolreg
Register input volumes to a base volume using AFNI 3dvolreg command
For complete details, see the 3dvolreg Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> volreg = afni.Volreg()
>>> volreg.inputs.in_file = 'functional.nii'
>>> volreg.inputs.args = '-Fourier -twopass'
>>> volreg.inputs.zpad = 4
>>> volreg.inputs.outputtype = "NIFTI"
>>> volreg.cmdline
'3dvolreg -Fourier -twopass -1Dfile functional.1D -1Dmatrix_save functional.aff12.1D -prefix functional_volreg.nii -zpad 4 -maxdisp1D functional_md.1D functional.nii'
>>> res = volreg.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dvolreg
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
basefile: (an existing file name)
base file for registration
flag: -base %s, position: -6
copyorigin: (a boolean)
copy base file origin coords to output
flag: -twodup
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
md1d_file: (a file name)
max displacement output file
flag: -maxdisp1D %s, position: -4
oned_file: (a file name)
1D movement parameters output file
flag: -1Dfile %s
oned_matrix_save: (a file name)
Save the matrix transformation
flag: -1Dmatrix_save %s
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
timeshift: (a boolean)
time shift to mean slice time offset
flag: -tshift 0
verbose: (a boolean)
more detailed description of the process
flag: -verbose
zpad: (an integer (int or long))
Zeropad around the edges by 'n' voxels during rotations
flag: -zpad %d, position: -5
Outputs:
md1d_file: (an existing file name)
max displacement info file
oned_file: (an existing file name)
movement parameters info file
oned_matrix_save: (an existing file name)
matrix transformation from base to input
out_file: (an existing file name)
registered file
Warp¶
Wraps command 3dWarp
Use 3dWarp for spatially transforming a dataset
For complete details, see the 3dWarp Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> warp = afni.Warp()
>>> warp.inputs.in_file = 'structural.nii'
>>> warp.inputs.deoblique = True
>>> warp.inputs.out_file = "trans.nii.gz"
>>> warp.cmdline
'3dWarp -deoblique -prefix trans.nii.gz structural.nii'
>>> warp_2 = afni.Warp()
>>> warp_2.inputs.in_file = 'structural.nii'
>>> warp_2.inputs.newgrid = 1.0
>>> warp_2.inputs.out_file = "trans.nii.gz"
>>> warp_2.cmdline
'3dWarp -newgrid 1.000000 -prefix trans.nii.gz structural.nii'
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dWarp
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
deoblique: (a boolean)
transform dataset from oblique to cardinal
flag: -deoblique
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
gridset: (an existing file name)
copy grid of specified dataset
flag: -gridset %s
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
interp: ('linear' or 'cubic' or 'NN' or 'quintic')
spatial interpolation methods [default = linear]
flag: -%s
matparent: (an existing file name)
apply transformation from 3dWarpDrive
flag: -matparent %s
mni2tta: (a boolean)
transform dataset from MNI152 to Talaraich
flag: -mni2tta
newgrid: (a float)
specify grid of this size (mm)
flag: -newgrid %f
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
tta2mni: (a boolean)
transform dataset from Talairach to MNI152
flag: -tta2mni
zpad: (an integer (int or long))
pad input dataset with N planes of zero on all sides.
flag: -zpad %d
Outputs:
out_file: (an existing file name)
output file
ZCutUp¶
Wraps command 3dZcutup
Cut z-slices from a volume using AFNI 3dZcutup command
For complete details, see the 3dZcutup Documentation.
Examples¶
>>> from nipype.interfaces import afni as afni
>>> zcutup = afni.ZCutUp()
>>> zcutup.inputs.in_file = 'functional.nii'
>>> zcutup.inputs.out_file = 'functional_zcutup.nii'
>>> zcutup.inputs.keep= '0 10'
>>> res = zcutup.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dZcutup
flag: %s, position: -1
[Optional]
args: (a string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a value of type 'str' and
with values which are a value of type 'str', nipype default value:
{})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
keep: (a string)
slice range to keep in output
flag: -keep %s
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('NIFTI_GZ' or 'AFNI' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file