interfaces.afni.utils¶
AFNItoNIFTI¶
Wraps command 3dAFNItoNIFTI
Converts AFNI format files to NIFTI format. This can also convert 2D or 1D data, which you can numpy.squeeze() to remove extra dimensions.
For complete details, see the 3dAFNItoNIFTI Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> a2n = afni.AFNItoNIFTI()
>>> a2n.inputs.in_file = 'afni_output.3D'
>>> a2n.inputs.out_file = 'afni_output.nii'
>>> a2n.cmdline
'3dAFNItoNIFTI -prefix afni_output.nii afni_output.3D'
>>> res = a2n.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dAFNItoNIFTI
flag: %s, position: -1
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
denote: (a boolean)
When writing the AFNI extension field, remove text notes that might
contain subject identifying information.
flag: -denote
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
newid: (a boolean)
Give the new dataset a new AFNI ID code, to distinguish it from the
input dataset.
flag: -newid
mutually_exclusive: oldid
oldid: (a boolean)
Give the new dataset the input datasets AFNI ID code.
flag: -oldid
mutually_exclusive: newid
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
pure: (a boolean)
Do NOT write an AFNI extension field into the output file. Only use
this option if needed. You can also use the 'nifti_tool' program to
strip extensions from a file.
flag: -pure
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
References:: None None
Autobox¶
Wraps command 3dAutobox
Computes size of a box that fits around the volume. Also can be used to crop the volume to that box.
For complete details, see the 3dAutobox Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> abox = afni.Autobox()
>>> abox.inputs.in_file = 'structural.nii'
>>> abox.inputs.padding = 5
>>> abox.cmdline
'3dAutobox -input structural.nii -prefix structural_autobox -npad 5'
>>> res = abox.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file
flag: -input %s
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
no_clustering: (a boolean)
Don't do any clustering to find box. Any non-zero voxel will be
preserved in the cropped volume. The default method uses some
clustering to find the cropping box, and will clip off small
isolated blobs.
flag: -noclust
out_file: (a file name)
flag: -prefix %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
padding: (an integer (int or long))
Number of extra voxels to pad on each side of box
flag: -npad %d
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (a file name)
output file
x_max: (an integer (int or long))
x_min: (an integer (int or long))
y_max: (an integer (int or long))
y_min: (an integer (int or long))
z_max: (an integer (int or long))
z_min: (an integer (int or long))
References:: None None
BrickStat¶
Wraps command 3dBrickStat
Computes maximum and/or minimum voxel values of an input dataset. TODO Add optional arguments.
For complete details, see the 3dBrickStat Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> brickstat = afni.BrickStat()
>>> brickstat.inputs.in_file = 'functional.nii'
>>> brickstat.inputs.mask = 'skeleton_mask.nii.gz'
>>> brickstat.inputs.min = True
>>> brickstat.cmdline
'3dBrickStat -min -mask skeleton_mask.nii.gz functional.nii'
>>> res = brickstat.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dmaskave
flag: %s, position: -1
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
-mask dset = use dset as mask to include/exclude voxels
flag: -mask %s, position: 2
min: (a boolean)
print the minimum value in dataset
flag: -min, position: 1
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
min_val: (a float)
output
Calc¶
Wraps command 3dcalc
This program does voxel-by-voxel arithmetic on 3D datasets.
For complete details, see the 3dcalc Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> calc = afni.Calc()
>>> calc.inputs.in_file_a = 'functional.nii'
>>> calc.inputs.in_file_b = 'functional2.nii'
>>> calc.inputs.expr='a*b'
>>> calc.inputs.out_file = 'functional_calc.nii.gz'
>>> calc.inputs.outputtype = 'NIFTI'
>>> calc.cmdline
'3dcalc -a functional.nii -b functional2.nii -expr "a*b" -prefix functional_calc.nii.gz'
>>> res = calc.run()
Inputs:
[Mandatory]
expr: (a unicode string)
expr
flag: -expr "%s", position: 3
in_file_a: (an existing file name)
input file to 3dcalc
flag: -a %s, position: 0
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
in_file_b: (an existing file name)
operand file to 3dcalc
flag: -b %s, position: 1
in_file_c: (an existing file name)
operand file to 3dcalc
flag: -c %s, position: 2
other: (a file name)
other options
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
single_idx: (an integer (int or long))
volume index for in_file_a
start_idx: (an integer (int or long))
start index for in_file_a
requires: stop_idx
stop_idx: (an integer (int or long))
stop index for in_file_a
requires: start_idx
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
References:: None None
Copy¶
Wraps command 3dcopy
Copies an image of one type to an image of the same or different type using 3dcopy command
For complete details, see the 3dcopy Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> copy3d = afni.Copy()
>>> copy3d.inputs.in_file = 'functional.nii'
>>> copy3d.cmdline
'3dcopy functional.nii functional_copy'
>>> res = copy3d.run()
>>> from copy import deepcopy
>>> copy3d_2 = deepcopy(copy3d)
>>> copy3d_2.inputs.outputtype = 'NIFTI'
>>> copy3d_2.cmdline
'3dcopy functional.nii functional_copy.nii'
>>> res = copy3d_2.run()
>>> copy3d_3 = deepcopy(copy3d)
>>> copy3d_3.inputs.outputtype = 'NIFTI_GZ'
>>> copy3d_3.cmdline
'3dcopy functional.nii functional_copy.nii.gz'
>>> res = copy3d_3.run()
>>> copy3d_4 = deepcopy(copy3d)
>>> copy3d_4.inputs.out_file = 'new_func.nii'
>>> copy3d_4.cmdline
'3dcopy functional.nii new_func.nii'
>>> res = copy3d_4.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dcopy
flag: %s, position: -2
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: %s, position: -1
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
References:: None None
Eval¶
Wraps command 1deval
Evaluates an expression that may include columns of data from one or more text files.
For complete details, see the 1deval Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> eval = afni.Eval()
>>> eval.inputs.in_file_a = 'seed.1D'
>>> eval.inputs.in_file_b = 'resp.1D'
>>> eval.inputs.expr = 'a*b'
>>> eval.inputs.out1D = True
>>> eval.inputs.out_file = 'data_calc.1D'
>>> eval.cmdline
'1deval -a seed.1D -b resp.1D -expr "a*b" -1D -prefix data_calc.1D'
>>> res = eval.run()
Inputs:
[Mandatory]
expr: (a unicode string)
expr
flag: -expr "%s", position: 3
in_file_a: (an existing file name)
input file to 1deval
flag: -a %s, position: 0
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
in_file_b: (an existing file name)
operand file to 1deval
flag: -b %s, position: 1
in_file_c: (an existing file name)
operand file to 1deval
flag: -c %s, position: 2
other: (a file name)
other options
out1D: (a boolean)
output in 1D
flag: -1D
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
single_idx: (an integer (int or long))
volume index for in_file_a
start_idx: (an integer (int or long))
start index for in_file_a
requires: stop_idx
stop_idx: (an integer (int or long))
stop index for in_file_a
requires: start_idx
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
References:: None None
FWHMx¶
Wraps command 3dFWHMx
Unlike the older 3dFWHM, this program computes FWHMs for all sub-bricks in the input dataset, each one separately. The output for each one is written to the file specified by ‘-out’. The mean (arithmetic or geometric) of all the FWHMs along each axis is written to stdout. (A non-positive output value indicates something bad happened; e.g., FWHM in z is meaningless for a 2D dataset; the estimation method computed incoherent intermediate results.)
For complete details, see the 3dFWHMx Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> fwhm = afni.FWHMx()
>>> fwhm.inputs.in_file = 'functional.nii'
>>> fwhm.cmdline
'3dFWHMx -input functional.nii -out functional_subbricks.out > functional_fwhmx.out'
>>> res = fwhm.run()
(Classic) METHOD:
- Calculate ratio of variance of first differences to data variance.
- Should be the same as 3dFWHM for a 1-brick dataset. (But the output format is simpler to use in a script.)
Note
IMPORTANT NOTE [AFNI > 16]
A completely new method for estimating and using noise smoothness values is now available in 3dFWHMx and 3dClustSim. This method is implemented in the ‘-acf’ options to both programs. ‘ACF’ stands for (spatial) AutoCorrelation Function, and it is estimated by calculating moments of differences out to a larger radius than before.
Notably, real FMRI data does not actually have a Gaussian-shaped ACF, so the estimated ACF is then fit (in 3dFWHMx) to a mixed model (Gaussian plus mono-exponential) of the form
where  is the radius, and
is the radius, and 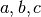 are the fitted parameters.
The apparent FWHM from this model is usually somewhat larger in real data
than the FWHM estimated from just the nearest-neighbor differences used
in the ‘classic’ analysis.
are the fitted parameters.
The apparent FWHM from this model is usually somewhat larger in real data
than the FWHM estimated from just the nearest-neighbor differences used
in the ‘classic’ analysis.
The longer tails provided by the mono-exponential are also significant. 3dClustSim has also been modified to use the ACF model given above to generate noise random fields.
Note
TL;DR or summary
The take-awaymessage is that the ‘classic’ 3dFWHMx and 3dClustSim analysis, using a pure Gaussian ACF, is not very correct for FMRI data – I cannot speak for PET or MEG data.
Warning
Do NOT use 3dFWHMx on the statistical results (e.g., ‘-bucket’) from 3dDeconvolve or 3dREMLfit!!! The function of 3dFWHMx is to estimate the smoothness of the time series NOISE, not of the statistics. This proscription is especially true if you plan to use 3dClustSim next!!
Note
Recommendations
- For FMRI statistical purposes, you DO NOT want the FWHM to reflect the spatial structure of the underlying anatomy. Rather, you want the FWHM to reflect the spatial structure of the noise. This means that the input dataset should not have anatomical (spatial) structure.
- One good form of input is the output of ‘3dDeconvolve -errts’, which is the dataset of residuals left over after the GLM fitted signal model is subtracted out from each voxel’s time series.
- If you don’t want to go to that much trouble, use ‘-detrend’ to approximately subtract out the anatomical spatial structure, OR use the output of 3dDetrend for the same purpose.
- If you do not use ‘-detrend’, the program attempts to find non-zero spatial structure in the input, and will print a warning message if it is detected.
Note
Notes on -demend
- I recommend this option, and it is not the default only for historical compatibility reasons. It may become the default someday.
- It is already the default in program 3dBlurToFWHM. This is the same detrending as done in 3dDespike; using 2*q+3 basis functions for q > 0.
- If you don’t use ‘-detrend’, the program now [Aug 2010] checks if a large number of voxels are have significant nonzero means. If so, the program will print a warning message suggesting the use of ‘-detrend’, since inherent spatial structure in the image will bias the estimation of the FWHM of the image time series NOISE (which is usually the point of using 3dFWHMx).
Inputs:
[Mandatory]
in_file: (an existing file name)
input dataset
flag: -input %s
[Optional]
acf: (a boolean or a file name or a tuple of the form: (an existing
file name, a float), nipype default value: False)
computes the spatial autocorrelation
flag: -acf
args: (a unicode string)
Additional parameters to the command
flag: %s
arith: (a boolean)
if in_file has more than one sub-brick, compute the final estimate
as the arithmetic mean of the individual sub-brick FWHM estimates
flag: -arith
mutually_exclusive: geom
automask: (a boolean, nipype default value: False)
compute a mask from THIS dataset, a la 3dAutomask
flag: -automask
combine: (a boolean)
combine the final measurements along each axis
flag: -combine
compat: (a boolean)
be compatible with the older 3dFWHM
flag: -compat
demed: (a boolean)
If the input dataset has more than one sub-brick (e.g., has a time
axis), then subtract the median of each voxel's time series before
processing FWHM. This will tend to remove intrinsic spatial
structure and leave behind the noise.
flag: -demed
mutually_exclusive: detrend
detrend: (a boolean or an integer (int or long), nipype default
value: False)
instead of demed (0th order detrending), detrend to the specified
order. If order is not given, the program picks q=NT/30. -detrend
disables -demed, and includes -unif.
flag: -detrend
mutually_exclusive: demed
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
geom: (a boolean)
if in_file has more than one sub-brick, compute the final estimate
as the geometric mean of the individual sub-brick FWHM estimates
flag: -geom
mutually_exclusive: arith
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
use only voxels that are nonzero in mask
flag: -mask %s
out_detrend: (a file name)
Save the detrended file into a dataset
flag: -detprefix %s
out_file: (a file name)
output file
flag: > %s, position: -1
out_subbricks: (a file name)
output file listing the subbricks FWHM
flag: -out %s
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
unif: (a boolean)
If the input dataset has more than one sub-brick, then normalize
each voxel's time series to have the same MAD before processing
FWHM.
flag: -unif
Outputs:
acf_param: (a tuple of the form: (a float, a float, a float) or a
tuple of the form: (a float, a float, a float, a float))
fitted ACF model parameters
fwhm: (a tuple of the form: (a float, a float, a float) or a tuple of
the form: (a float, a float, a float, a float))
FWHM along each axis
out_acf: (an existing file name)
output acf file
out_detrend: (a file name)
output file, detrended
out_file: (an existing file name)
output file
out_subbricks: (an existing file name)
output file (subbricks)
References:: None
MaskTool¶
Wraps command 3dmask_tool
3dmask_tool - for combining/dilating/eroding/filling masks
For complete details, see the 3dmask_tool Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> masktool = afni.MaskTool()
>>> masktool.inputs.in_file = 'functional.nii'
>>> masktool.inputs.outputtype = 'NIFTI'
>>> masktool.cmdline
'3dmask_tool -prefix functional_mask.nii -input functional.nii'
>>> res = automask.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file or files to 3dmask_tool
flag: -input %s, position: -1
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
count: (a boolean)
Instead of created a binary 0/1 mask dataset, create one with counts
of voxel overlap, i.e., each voxel will contain the number of masks
that it is set in.
flag: -count, position: 2
datum: ('byte' or 'short' or 'float')
specify data type for output. Valid types are 'byte', 'short' and
'float'.
flag: -datum %s
dilate_inputs: (a unicode string)
Use this option to dilate and/or erode datasets as they are read.
ex. '5 -5' to dilate and erode 5 times
flag: -dilate_inputs %s
dilate_results: (a unicode string)
dilate and/or erode combined mask at the given levels.
flag: -dilate_results %s
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
fill_dirs: (a unicode string)
fill holes only in the given directions. This option is for use with
-fill holes. should be a single string that specifies 1-3 of the
axes using {x,y,z} labels (i.e. dataset axis order), or using the
labels in {R,L,A,P,I,S}.
flag: -fill_dirs %s
requires: fill_holes
fill_holes: (a boolean)
This option can be used to fill holes in the resulting mask, i.e.
after all other processing has been done.
flag: -fill_holes
frac: (a float)
When combining masks (across datasets and sub-bricks), use this
option to restrict the result to a certain fraction of the set of
volumes
flag: -frac %s
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
inter: (a boolean)
intersection, this means -frac 1.0
flag: -inter
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
union: (a boolean)
union, this means -frac 0
flag: -union
Outputs:
out_file: (an existing file name)
mask file
References:: None None
Merge¶
Wraps command 3dmerge
Merge or edit volumes using AFNI 3dmerge command
For complete details, see the 3dmerge Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> merge = afni.Merge()
>>> merge.inputs.in_files = ['functional.nii', 'functional2.nii']
>>> merge.inputs.blurfwhm = 4
>>> merge.inputs.doall = True
>>> merge.inputs.out_file = 'e7.nii'
>>> merge.cmdline
'3dmerge -1blur_fwhm 4 -doall -prefix e7.nii functional.nii functional2.nii'
>>> res = merge.run()
Inputs:
[Mandatory]
in_files: (a list of items which are an existing file name)
flag: %s, position: -1
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
blurfwhm: (an integer (int or long))
FWHM blur value (mm)
flag: -1blur_fwhm %d
doall: (a boolean)
apply options to all sub-bricks in dataset
flag: -doall
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
References:: None None
Notes¶
Wraps command 3dNotes
A program to add, delete, and show notes for AFNI datasets.
For complete details, see the 3dNotes Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> notes = afni.Notes()
>>> notes.inputs.in_file = 'functional.HEAD'
>>> notes.inputs.add = 'This note is added.'
>>> notes.inputs.add_history = 'This note is added to history.'
>>> notes.cmdline
'3dNotes -a "This note is added." -h "This note is added to history." functional.HEAD'
>>> res = notes.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dNotes
flag: %s, position: -1
[Optional]
add: (a unicode string)
note to add
flag: -a "%s"
add_history: (a unicode string)
note to add to history
flag: -h "%s"
mutually_exclusive: rep_history
args: (a unicode string)
Additional parameters to the command
flag: %s
delete: (an integer (int or long))
delete note number num
flag: -d %d
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
rep_history: (a unicode string)
note with which to replace history
flag: -HH "%s"
mutually_exclusive: add_history
ses: (a boolean)
print to stdout the expanded notes
flag: -ses
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
Refit¶
Wraps command 3drefit
Changes some of the information inside a 3D dataset’s header
For complete details, see the 3drefit Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> refit = afni.Refit()
>>> refit.inputs.in_file = 'structural.nii'
>>> refit.inputs.deoblique = True
>>> refit.cmdline
'3drefit -deoblique structural.nii'
>>> res = refit.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3drefit
flag: %s, position: -1
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
deoblique: (a boolean)
replace current transformation matrix with cardinal matrix
flag: -deoblique
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
space: ('TLRC' or 'MNI' or 'ORIG')
Associates the dataset with a specific template type, e.g. TLRC,
MNI, ORIG
flag: -space %s
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
xdel: (a float)
new x voxel dimension in mm
flag: -xdel %f
xorigin: (a unicode string)
x distance for edge voxel offset
flag: -xorigin %s
ydel: (a float)
new y voxel dimension in mm
flag: -ydel %f
yorigin: (a unicode string)
y distance for edge voxel offset
flag: -yorigin %s
zdel: (a float)
new z voxel dimension in mm
flag: -zdel %f
zorigin: (a unicode string)
z distance for edge voxel offset
flag: -zorigin %s
Outputs:
out_file: (an existing file name)
output file
Resample¶
Wraps command 3dresample
Resample or reorient an image using AFNI 3dresample command
For complete details, see the 3dresample Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> resample = afni.Resample()
>>> resample.inputs.in_file = 'functional.nii'
>>> resample.inputs.orientation= 'RPI'
>>> resample.inputs.outputtype = 'NIFTI'
>>> resample.cmdline
'3dresample -orient RPI -prefix functional_resample.nii -inset functional.nii'
>>> res = resample.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dresample
flag: -inset %s, position: -1
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
master: (a file name)
align dataset grid to a reference file
flag: -master %s
orientation: (a unicode string)
new orientation code
flag: -orient %s
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
resample_mode: ('NN' or 'Li' or 'Cu' or 'Bk')
resampling method from set {"NN", "Li", "Cu", "Bk"}. These are for
"Nearest Neighbor", "Linear", "Cubic" and "Blocky"interpolation,
respectively. Default is NN.
flag: -rmode %s
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
voxel_size: (a tuple of the form: (a float, a float, a float))
resample to new dx, dy and dz
flag: -dxyz %f %f %f
Outputs:
out_file: (an existing file name)
output file
References:: None None
TCat¶
Wraps command 3dTcat
Concatenate sub-bricks from input datasets into one big 3D+time dataset.
TODO Replace InputMultiPath in_files with Traits.List, if possible. Current version adds extra whitespace.
For complete details, see the 3dTcat Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> tcat = afni.TCat()
>>> tcat.inputs.in_files = ['functional.nii', 'functional2.nii']
>>> tcat.inputs.out_file= 'functional_tcat.nii'
>>> tcat.inputs.rlt = '+'
>>> tcat.cmdline
'3dTcat -rlt+ -prefix functional_tcat.nii functional.nii functional2.nii'
>>> res = tcat.run()
Inputs:
[Mandatory]
in_files: (a list of items which are an existing file name)
input file to 3dTcat
flag: %s, position: -1
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
rlt: ('' or '+' or '++')
Remove linear trends in each voxel time series loaded from each
input dataset, SEPARATELY. Option -rlt removes the least squares fit
of 'a+b*t' to each voxel time series. Option -rlt+ adds dataset mean
back in. Option -rlt++ adds overall mean of all dataset timeseries
back in.
flag: -rlt%s, position: 1
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
References:: None None
TStat¶
Wraps command 3dTstat
Compute voxel-wise statistics using AFNI 3dTstat command
For complete details, see the 3dTstat Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> tstat = afni.TStat()
>>> tstat.inputs.in_file = 'functional.nii'
>>> tstat.inputs.args = '-mean'
>>> tstat.inputs.out_file = 'stats'
>>> tstat.cmdline
'3dTstat -mean -prefix stats functional.nii'
>>> res = tstat.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dTstat
flag: %s, position: -1
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
mask: (an existing file name)
mask file
flag: -mask %s
options: (a unicode string)
selected statistical output
flag: %s
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
References:: None None
To3D¶
Wraps command to3d
Create a 3D dataset from 2D image files using AFNI to3d command
For complete details, see the to3d Documentation
Examples¶
>>> from nipype.interfaces import afni
>>> to3d = afni.To3D()
>>> to3d.inputs.datatype = 'float'
>>> to3d.inputs.in_folder = '.'
>>> to3d.inputs.out_file = 'dicomdir.nii'
>>> to3d.inputs.filetype = 'anat'
>>> to3d.cmdline
'to3d -datum float -anat -prefix dicomdir.nii ./*.dcm'
>>> res = to3d.run()
Inputs:
[Mandatory]
in_folder: (an existing directory name)
folder with DICOM images to convert
flag: %s/*.dcm, position: -1
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
assumemosaic: (a boolean)
assume that Siemens image is mosaic
flag: -assume_dicom_mosaic
datatype: ('short' or 'float' or 'byte' or 'complex')
set output file datatype
flag: -datum %s
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
filetype: ('spgr' or 'fse' or 'epan' or 'anat' or 'ct' or 'spct' or
'pet' or 'mra' or 'bmap' or 'diff' or 'omri' or 'abuc' or 'fim' or
'fith' or 'fico' or 'fitt' or 'fift' or 'fizt' or 'fict' or 'fibt'
or 'fibn' or 'figt' or 'fipt' or 'fbuc')
type of datafile being converted
flag: -%s
funcparams: (a unicode string)
parameters for functional data
flag: -time:zt %s alt+z2
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
skipoutliers: (a boolean)
skip the outliers check
flag: -skip_outliers
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
References:: None None
Unifize¶
Wraps command 3dUnifize
3dUnifize - for uniformizing image intensity
- The input dataset is supposed to be a T1-weighted volume, possibly already skull-stripped (e.g., via 3dSkullStrip). However, this program can be a useful step to take BEFORE 3dSkullStrip, since the latter program can fail if the input volume is strongly shaded – 3dUnifize will (mostly) remove such shading artifacts.
- The output dataset has the white matter (WM) intensity approximately uniformized across space, and scaled to peak at about 1000.
- The output dataset is always stored in float format!
- If the input dataset has more than 1 sub-brick, only sub-brick #0 will be processed!
- Want to correct EPI datasets for nonuniformity? You can try the new and experimental [Mar 2017] ‘-EPI’ option.
- The principal motive for this program is for use in an image registration script, and it may or may not be useful otherwise.
- This program replaces the older (and very different) 3dUniformize, which is no longer maintained and may sublimate at any moment. (In other words, we do not recommend the use of 3dUniformize.)
For complete details, see the 3dUnifize Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> unifize = afni.Unifize()
>>> unifize.inputs.in_file = 'structural.nii'
>>> unifize.inputs.out_file = 'structural_unifized.nii'
>>> unifize.cmdline
'3dUnifize -prefix structural_unifized.nii -input structural.nii'
>>> res = unifize.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dUnifize
flag: -input %s, position: -1
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
epi: (a boolean)
Assume the input dataset is a T2 (or T2*) weighted EPI time series.
After computing the scaling, apply it to ALL volumes (TRs) in the
input dataset. That is, a given voxel will be scaled by the same
factor at each TR. This option also implies '-noduplo' and
'-T2'.This option turns off '-GM' if you turned it on.
flag: -EPI
mutually_exclusive: gm
requires: no_duplo, t2
gm: (a boolean)
Also scale to unifize 'gray matter' = lower intensity voxels (to aid
in registering images from different scanners).
flag: -GM
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
no_duplo: (a boolean)
Do NOT use the 'duplo down' step; this can be useful for lower
resolution datasets.
flag: -noduplo
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
scale_file: (a file name)
output file name to save the scale factor used at each voxel
flag: -ssave %s
t2: (a boolean)
Treat the input as if it were T2-weighted, rather than T1-weighted.
This processing is done simply by inverting the image contrast,
processing it as if that result were T1-weighted, and then re-
inverting the results counts of voxel overlap, i.e., each voxel will
contain the number of masks that it is set in.
flag: -T2
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
urad: (a float)
Sets the radius (in voxels) of the ball used for the sneaky trick.
Default value is 18.3, and should be changed proportionally if the
dataset voxel size differs significantly from 1 mm.
flag: -Urad %s
Outputs:
out_file: (an existing file name)
unifized file
scale_file: (a file name)
scale factor file
References:: None None
ZCutUp¶
Wraps command 3dZcutup
Cut z-slices from a volume using AFNI 3dZcutup command
For complete details, see the 3dZcutup Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> zcutup = afni.ZCutUp()
>>> zcutup.inputs.in_file = 'functional.nii'
>>> zcutup.inputs.out_file = 'functional_zcutup.nii'
>>> zcutup.inputs.keep= '0 10'
>>> zcutup.cmdline
'3dZcutup -keep 0 10 -prefix functional_zcutup.nii functional.nii'
>>> res = zcutup.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dZcutup
flag: %s, position: -1
[Optional]
args: (a unicode string)
Additional parameters to the command
flag: %s
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a value
of class 'str', nipype default value: {})
Environment variables
ignore_exception: (a boolean, nipype default value: False)
Print an error message instead of throwing an exception in case the
interface fails to run
keep: (a unicode string)
slice range to keep in output
flag: -keep %s
out_file: (a file name)
output image file name
flag: -prefix %s
outputtype: ('AFNI' or 'NIFTI_GZ' or 'NIFTI')
AFNI output filetype
terminal_output: ('stream' or 'allatonce' or 'file' or 'none')
Control terminal output: `stream` - displays to terminal immediately
(default), `allatonce` - waits till command is finished to display
output, `file` - writes output to file, `none` - output is ignored
Outputs:
out_file: (an existing file name)
output file
References:: None None