Pipeline 102¶
Now that you know how to construct a workflow and execute it, we will go
into more advanced concepts. This tutorial focuses on
nipype.pipeline.engine.Workflow
nipype.pipeline.engine.Node and
nipype.pipeline.engine.MapNode.
A workflow is a directed acyclic graph (DAG) consisting of nodes which can be of type Workflow, Node or MapNode. Workflows can be re-used and hierarchical workflows can be easily constructed.
‘name’ : the mandatory keyword arg¶
When instantiating a Workflow, Node or MapNode, a name has to be provided. For any given level of a workflow, no two nodes can have the same name. The engine will let you know if this is the case when you add nodes to a workflow either directly using add_nodes or using the connect function.
Names have many internal uses. They determine the name of the directory in which the workflow/node is run and the outputs are stored.
realigner = pe.Node(interface=spm.Realign(),
name='RealignSPM')
Now this output will be stored in a directory called RealignSPM. Proper naming of your nodes can be advantageous from the perspective that it provides a semantic descriptor aligned with your thought process. This name parameter is also used to refer to nodes in embedded workflows.
iterables¶
This can only be set for Node and MapNode. This is syntactic sugar for
running a subgraph with the Node/MapNode at its root in a for
loop. For example, consider an fMRI preprocessing pipeline that you
would like to run for all your subjects. You can define a workflow and
then execute it for every single subject inside a for loop. Consider
the simplistic example below, where startnode is a node belonging to
workflow ‘mywork.’
for s in subjects:
startnode.inputs.subject_id = s
mywork.run()
The pipeline engine provides a convenience function that simplifies this:
startnode.iterables = ('subject_id', subjects)
mywork.run()
This will achieve the same exact behavior as the for loop above. The workflow graph is:
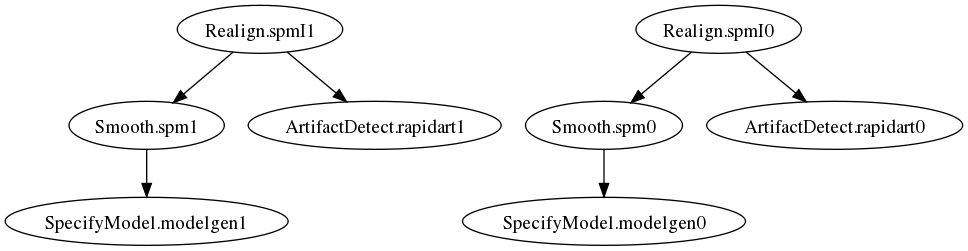
Now consider the situation in which you want the last node (typically smoothing) of your preprocessing pipeline to smooth using two different kernels (0 mm and 6 mm FWHM). Again the common approach would be:
for s in subjects:
startnode.inputs.subject_id = s
uptosmoothingworkflow.run()
smoothnode.inputs.infile = lastnode.output.outfile
for fwhm in [0, 6]:
smoothnode.inputs.fwhm = fwhm
remainingworkflow.run()
Instead of having multiple for loops at various stages, you can set up
another set of iterables for the smoothnode.
startnode.iterables = ('subject_id', subjects)
smoothnode.iterables = ('fwhm', [0, 6])
mywork.run()
This will run the preprocessing workflow for two different smoothing kernels over all subjects.
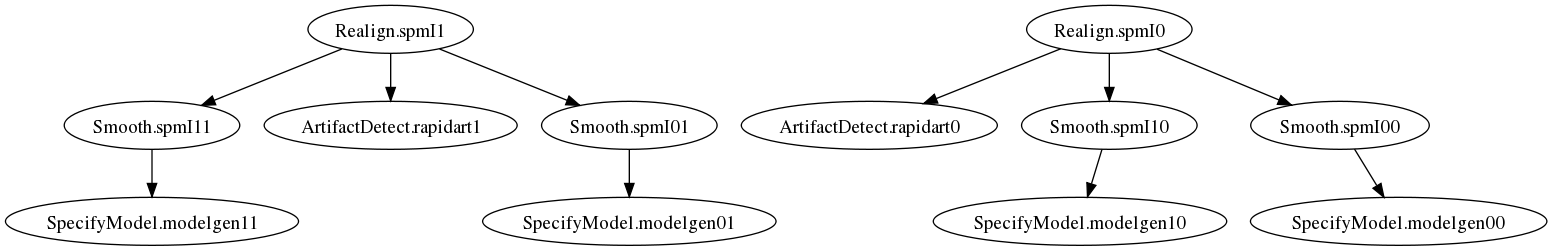
Thus setting iterables has a multiplicative effect. In the above examples there is a separate, distinct specifymodel node that’s executed for each combination of subject and smoothing.
iterfield¶
This is a mandatory keyword arg for MapNode. This enables running the
underlying interface over a set of inputs and is particularly useful
when the interface can only operate on a single input. For example, the
nipype.interfaces.fsl.BET will operate on only one (3d or 4d)
NIfTI file. But wrapping BET in a MapNode can execute it over a list of files:
better = pe.MapNode(interface=fsl.Bet(), name='stripper',
iterfield=['in_file'])
better.inputs.in_file = ['file1.nii','file2.nii']
better.run()
This will create a directory called stripper and inside it two
subdirectories called in_file_0 and in_file_1. The output of running bet
separately on each of those files will be stored in those two
subdirectories.
This can be extended to run it on pairwise inputs. For example,
transform = pe.MapNode(interface=fs.ApplyVolTransform(),
name='warpvol',
iterfield=['source_file', 'reg_file'])
transform.inputs.source_file = ['file1.nii','file2.nii']
transform.inputs.reg_file = ['file1.reg','file2.reg']
transform.run()
The above will be equivalent to running transform by taking corresponding items from each of the two fields in iterfield. The subdirectories get always named with respect to the first iterfield.
overwrite¶
The overwrite keyword arg forces a node to be rerun.
The clone function¶
The clone function can be used to create a copy of a workflow. No references to the original workflow are retained. As such the clone function requires a name keyword arg that specifies a new name for the duplicate workflow.





