interfaces.afni.utils¶
ABoverlap¶
Wraps the executable command 3dABoverlap.
Output (to screen) is a count of various things about how the automasks of datasets A and B overlap or don’t overlap.
For complete details, see the 3dABoverlap Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> aboverlap = afni.ABoverlap()
>>> aboverlap.inputs.in_file_a = 'functional.nii'
>>> aboverlap.inputs.in_file_b = 'structural.nii'
>>> aboverlap.inputs.out_file = 'out.mask_ae_overlap.txt'
>>> aboverlap.cmdline
'3dABoverlap functional.nii structural.nii |& tee out.mask_ae_overlap.txt'
>>> res = aboverlap.run()
Inputs:
[Mandatory]
in_file_a: (an existing file name)
input file A
argument: ``%s``, position: -3
in_file_b: (an existing file name)
input file B
argument: ``%s``, position: -2
[Optional]
out_file: (a file name)
collect output to a file
argument: `` |& tee %s``, position: -1
no_automask: (a boolean)
consider input datasets as masks
argument: ``-no_automask``
quiet: (a boolean)
be as quiet as possible (without being entirely mute)
argument: ``-quiet``
verb: (a boolean)
print out some progress reports (to stderr)
argument: ``-verb``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
AFNItoNIFTI¶
Wraps the executable command 3dAFNItoNIFTI.
Converts AFNI format files to NIFTI format. This can also convert 2D or 1D data, which you can numpy.squeeze() to remove extra dimensions.
For complete details, see the 3dAFNItoNIFTI Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> a2n = afni.AFNItoNIFTI()
>>> a2n.inputs.in_file = 'afni_output.3D'
>>> a2n.inputs.out_file = 'afni_output.nii'
>>> a2n.cmdline
'3dAFNItoNIFTI -prefix afni_output.nii afni_output.3D'
>>> res = a2n.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dAFNItoNIFTI
argument: ``%s``, position: -1
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``
pure: (a boolean)
Do NOT write an AFNI extension field into the output file. Only use
this option if needed. You can also use the 'nifti_tool' program to
strip extensions from a file.
argument: ``-pure``
denote: (a boolean)
When writing the AFNI extension field, remove text notes that might
contain subject identifying information.
argument: ``-denote``
oldid: (a boolean)
Give the new dataset the input datasets AFNI ID code.
argument: ``-oldid``
mutually_exclusive: newid
newid: (a boolean)
Give the new dataset a new AFNI ID code, to distinguish it from the
input dataset.
argument: ``-newid``
mutually_exclusive: oldid
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
Autobox¶
Wraps the executable command 3dAutobox.
Computes size of a box that fits around the volume. Also can be used to crop the volume to that box.
For complete details, see the 3dAutobox Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> abox = afni.Autobox()
>>> abox.inputs.in_file = 'structural.nii'
>>> abox.inputs.padding = 5
>>> abox.cmdline
'3dAutobox -input structural.nii -prefix structural_autobox -npad 5'
>>> res = abox.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file
argument: ``-input %s``
[Optional]
padding: (an integer (int or long))
Number of extra voxels to pad on each side of box
argument: ``-npad %d``
out_file: (a file name)
argument: ``-prefix %s``
no_clustering: (a boolean)
Don't do any clustering to find box. Any non-zero voxel will be
preserved in the cropped volume. The default method uses some
clustering to find the cropping box, and will clip off small
isolated blobs.
argument: ``-noclust``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
x_min: (an integer (int or long))
x_max: (an integer (int or long))
y_min: (an integer (int or long))
y_max: (an integer (int or long))
z_min: (an integer (int or long))
z_max: (an integer (int or long))
out_file: (a file name)
output file
References:¶
None None
Axialize¶
Wraps the executable command 3daxialize.
- Read in a dataset and write it out as a new dataset
- with the data brick oriented as axial slices.
For complete details, see the 3dcopy Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> axial3d = afni.Axialize()
>>> axial3d.inputs.in_file = 'functional.nii'
>>> axial3d.inputs.out_file = 'axialized.nii'
>>> axial3d.cmdline
'3daxialize -prefix axialized.nii functional.nii'
>>> res = axial3d.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3daxialize
argument: ``%s``, position: -2
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``
verb: (a boolean)
Print out a progerss report
argument: ``-verb``
sagittal: (a boolean)
Do sagittal slice order [-orient ASL]
argument: ``-sagittal``
mutually_exclusive: coronal, axial
coronal: (a boolean)
Do coronal slice order [-orient RSA]
argument: ``-coronal``
mutually_exclusive: sagittal, axial
axial: (a boolean)
Do axial slice order [-orient RAI]This is the default AFNI axial
order, andis the one currently required by thevolume rendering
plugin; this is alsothe default orientation output by thisprogram
(hence the program's name).
argument: ``-axial``
mutually_exclusive: coronal, sagittal
orientation: (a unicode string)
new orientation code
argument: ``-orient %s``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
BrickStat¶
Wraps the executable command 3dBrickStat.
Computes maximum and/or minimum voxel values of an input dataset. TODO Add optional arguments.
For complete details, see the 3dBrickStat Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> brickstat = afni.BrickStat()
>>> brickstat.inputs.in_file = 'functional.nii'
>>> brickstat.inputs.mask = 'skeleton_mask.nii.gz'
>>> brickstat.inputs.min = True
>>> brickstat.cmdline
'3dBrickStat -min -mask skeleton_mask.nii.gz functional.nii'
>>> res = brickstat.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dmaskave
argument: ``%s``, position: -1
[Optional]
mask: (an existing file name)
-mask dset = use dset as mask to include/exclude voxels
argument: ``-mask %s``, position: 2
min: (a boolean)
print the minimum value in dataset
argument: ``-min``, position: 1
slow: (a boolean)
read the whole dataset to find the min and max values
argument: ``-slow``
max: (a boolean)
print the maximum value in the dataset
argument: ``-max``
mean: (a boolean)
print the mean value in the dataset
argument: ``-mean``
sum: (a boolean)
print the sum of values in the dataset
argument: ``-sum``
var: (a boolean)
print the variance in the dataset
argument: ``-var``
percentile: (a tuple of the form: (a float, a float, a float))
p0 ps p1 write the percentile values starting at p0% and ending at
p1% at a step of ps%. only one sub-brick is accepted.
argument: ``-percentile %.3f %.3f %.3f``
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
min_val: (a float)
output
Bucket¶
Wraps the executable command 3dbucket.
Concatenate sub-bricks from input datasets into one big ‘bucket’ dataset.
For complete details, see the 3dbucket Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> bucket = afni.Bucket()
>>> bucket.inputs.in_file = [('functional.nii',"{2..$}"), ('functional.nii',"{1}")]
>>> bucket.inputs.out_file = 'vr_base'
>>> bucket.cmdline
"3dbucket -prefix vr_base functional.nii'{2..$}' functional.nii'{1}'"
>>> res = bucket.run()
Inputs:
[Mandatory]
in_file: (a list of items which are a tuple of the form: (an existing
file name, a unicode string))
List of tuples of input datasets and subbrick selection stringsas
described in more detail in the following afni help stringInput
dataset specified using one of these forms: 'prefix+view',
'prefix+view.HEAD', or 'prefix+view.BRIK'.You can also add a sub-
brick selection list after the end of thedataset name. This allows
only a subset of the sub-bricks to beincluded into the output (by
default, all of the input datasetis copied into the output). A sub-
brick selection list looks likeone of the following forms:
fred+orig[5] ==> use only sub-brick #5 fred+orig[5,9,17] ==> use #5,
#9, and #17 fred+orig[5..8] or [5-8] ==> use #5, #6, #7, and #8
fred+orig[5..13(2)] or [5-13(2)] ==> use #5, #7, #9, #11, and
#13Sub-brick indexes start at 0. You can use the character '$'to
indicate the last sub-brick in a dataset; for example, youcan select
every third sub-brick by using the selection list
fred+orig[0..$(3)]N.B.: The sub-bricks are output in the order
specified, which may not be the order in the original datasets. For
example, using fred+orig[0..$(2),1..$(2)] will cause the sub-bricks
in fred+orig to be output into the new dataset in an interleaved
fashion. Using fred+orig[$..0] will reverse the order of the sub-
bricks in the output.N.B.: Bucket datasets have multiple sub-bricks,
but do NOT have a time dimension. You can input sub-bricks from a
3D+time dataset into a bucket dataset. You can use the '3dinfo'
program to see how many sub-bricks a 3D+time or a bucket dataset
contains.N.B.: In non-bucket functional datasets (like the 'fico'
datasets output by FIM, or the 'fitt' datasets output by 3dttest),
sub-brick [0] is the 'intensity' and sub-brick [1] is the
statistical parameter used as a threshold. Thus, to create a bucket
dataset using the intensity from dataset A and the threshold from
dataset B, and calling the output dataset C, you would type 3dbucket
-prefix C -fbuc 'A+orig[0]' -fbuc 'B+orig[1]'WARNING: using this
program, it is possible to create a dataset that has different basic
datum types for different sub-bricks (e.g., shorts for brick 0,
floats for brick 1). Do NOT do this! Very few AFNI programs will
work correctly with such datasets!
argument: ``%s``, position: -1
[Optional]
out_file: (a file name)
argument: ``-prefix %s``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
Calc¶
Wraps the executable command 3dcalc.
This program does voxel-by-voxel arithmetic on 3D datasets.
For complete details, see the 3dcalc Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> calc = afni.Calc()
>>> calc.inputs.in_file_a = 'functional.nii'
>>> calc.inputs.in_file_b = 'functional2.nii'
>>> calc.inputs.expr='a*b'
>>> calc.inputs.out_file = 'functional_calc.nii.gz'
>>> calc.inputs.outputtype = 'NIFTI'
>>> calc.cmdline
'3dcalc -a functional.nii -b functional2.nii -expr "a*b" -prefix functional_calc.nii.gz'
>>> res = calc.run()
>>> from nipype.interfaces import afni
>>> calc = afni.Calc()
>>> calc.inputs.in_file_a = 'functional.nii'
>>> calc.inputs.expr = '1'
>>> calc.inputs.out_file = 'rm.epi.all1'
>>> calc.inputs.overwrite = True
>>> calc.cmdline
'3dcalc -a functional.nii -expr "1" -prefix rm.epi.all1 -overwrite'
>>> res = calc.run()
Inputs:
[Mandatory]
in_file_a: (an existing file name)
input file to 3dcalc
argument: ``-a %s``, position: 0
expr: (a unicode string)
expr
argument: ``-expr "%s"``, position: 3
[Optional]
in_file_b: (an existing file name)
operand file to 3dcalc
argument: ``-b %s``, position: 1
in_file_c: (an existing file name)
operand file to 3dcalc
argument: ``-c %s``, position: 2
out_file: (a file name)
output image file name
argument: ``-prefix %s``
start_idx: (an integer (int or long))
start index for in_file_a
requires: stop_idx
stop_idx: (an integer (int or long))
stop index for in_file_a
requires: start_idx
single_idx: (an integer (int or long))
volume index for in_file_a
overwrite: (a boolean)
overwrite output
argument: ``-overwrite``
other: (a file name)
other options
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
Cat¶
Wraps the executable command 1dcat.
1dcat takes as input one or more 1D files, and writes out a 1D file containing the side-by-side concatenation of all or a subset of the columns from the input files.
For complete details, see the 1dcat Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> cat1d = afni.Cat()
>>> cat1d.inputs.sel = "'[0,2]'"
>>> cat1d.inputs.in_files = ['f1.1D', 'f2.1D']
>>> cat1d.inputs.out_file = 'catout.1d'
>>> cat1d.cmdline
"1dcat -sel '[0,2]' f1.1D f2.1D > catout.1d"
>>> res = cat1d.run()
Inputs:
[Mandatory]
in_files: (a list of items which are an existing file name)
argument: ``%s``, position: -2
out_file: (a file name, nipype default value: catout.1d)
output (concatenated) file name
argument: ``> %s``, position: -1
[Optional]
omitconst: (a boolean)
Omit columns that are identically constant from output.
argument: ``-nonconst``
keepfree: (a boolean)
Keep only columns that are marked as 'free' in the 3dAllineate
header from '-1Dparam_save'. If there is no such header, all columns
are kept.
argument: ``-nonfixed``
out_format: ('int' or 'nice' or 'double' or 'fint' or 'cint')
specify data type for output. Valid types are 'int', 'nice',
'double', 'fint', and 'cint'.
argument: ``-form %s``
mutually_exclusive: out_int, out_nice, out_double, out_fint,
out_cint
stack: (a boolean)
Stack the columns of the resultant matrix in the output.
argument: ``-stack``
sel: (a unicode string)
Apply the same column/row selection string to all filenames on the
command line.
argument: ``-sel %s``
out_int: (a boolean)
specifiy int data type for output
argument: ``-i``
mutually_exclusive: out_format, out_nice, out_double, out_fint,
out_cint
out_nice: (a boolean)
specifiy nice data type for output
argument: ``-n``
mutually_exclusive: out_format, out_int, out_double, out_fint,
out_cint
out_double: (a boolean)
specifiy double data type for output
argument: ``-d``
mutually_exclusive: out_format, out_nice, out_int, out_fint,
out_cint
out_fint: (a boolean)
specifiy int, rounded down, data type for output
argument: ``-f``
mutually_exclusive: out_format, out_nice, out_double, out_int,
out_cint
out_cint: (a boolean)
specifiy int, rounded up, data type for output
mutually_exclusive: out_format, out_nice, out_double, out_fint,
out_int
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
CatMatvec¶
Wraps the executable command cat_matvec.
Catenates 3D rotation+shift matrix+vector transformations.
For complete details, see the cat_matvec Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> cmv = afni.CatMatvec()
>>> cmv.inputs.in_file = [('structural.BRIK::WARP_DATA','I')]
>>> cmv.inputs.out_file = 'warp.anat.Xat.1D'
>>> cmv.cmdline
'cat_matvec structural.BRIK::WARP_DATA -I > warp.anat.Xat.1D'
>>> res = cmv.run()
Inputs:
[Mandatory]
in_file: (a list of items which are a tuple of the form: (a unicode
string, a unicode string))
list of tuples of mfiles and associated opkeys
argument: ``%s``, position: -2
out_file: (a file name)
File to write concattenated matvecs to
argument: `` > %s``, position: -1
[Optional]
matrix: (a boolean)
indicates that the resulting matrix willbe written to outfile in the
'MATRIX(...)' format (FORM 3).This feature could be used, with
clever scripting, to inputa matrix directly on the command line to
program 3dWarp.
argument: ``-MATRIX``
mutually_exclusive: oneline, fourxfour
oneline: (a boolean)
indicates that the resulting matrixwill simply be written as 12
numbers on one line.
argument: ``-ONELINE``
mutually_exclusive: matrix, fourxfour
fourxfour: (a boolean)
Output matrix in augmented form (last row is 0 0 0 1)This option
does not work with -MATRIX or -ONELINE
argument: ``-4x4``
mutually_exclusive: matrix, oneline
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
CenterMass¶
Wraps the executable command 3dCM.
Computes center of mass using 3dCM command
Note
By default, the output is (x,y,z) values in DICOM coordinates. But as of Dec, 2016, there are now command line switches for other options.
For complete details, see the 3dCM Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> cm = afni.CenterMass()
>>> cm.inputs.in_file = 'structural.nii'
>>> cm.inputs.cm_file = 'cm.txt'
>>> cm.inputs.roi_vals = [2, 10]
>>> cm.cmdline
'3dCM -roi_vals 2 10 structural.nii > cm.txt'
>>> res = 3dcm.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dCM
argument: ``%s``, position: -2
[Optional]
cm_file: (a file name)
File to write center of mass to
argument: ``> %s``, position: -1
mask_file: (an existing file name)
Only voxels with nonzero values in the provided mask will be
averaged.
argument: ``-mask %s``
automask: (a boolean)
Generate the mask automatically
argument: ``-automask``
set_cm: (a tuple of the form: (a float, a float, a float))
After computing the center of mass, set the origin fields in the
header so that the center of mass will be at (x,y,z) in DICOM
coords.
argument: ``-set %f %f %f``
local_ijk: (a boolean)
Output values as (i,j,k) in local orienation
argument: ``-local_ijk``
roi_vals: (a list of items which are an integer (int or long))
Compute center of mass for each blob with voxel value of v0, v1, v2,
etc. This option is handy for getting ROI centers of mass.
argument: ``-roi_vals %s``
all_rois: (a boolean)
Don't bother listing the values of ROIs you want: The program will
find all of them and produce a full list
argument: ``-all_rois``
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
cm_file: (a file name)
file with the center of mass coordinates
cm: (a list of items which are a tuple of the form: (a float, a
float, a float))
center of mass
ConvertDset¶
Wraps the executable command ConvertDset.
Converts a surface dataset from one format to another.
For complete details, see the ConvertDset Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> convertdset = afni.ConvertDset()
>>> convertdset.inputs.in_file = 'lh.pial_converted.gii'
>>> convertdset.inputs.out_type = 'niml_asc'
>>> convertdset.inputs.out_file = 'lh.pial_converted.niml.dset'
>>> convertdset.cmdline
'ConvertDset -o_niml_asc -input lh.pial_converted.gii -prefix lh.pial_converted.niml.dset'
>>> res = convertdset.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to ConvertDset
argument: ``-input %s``, position: -2
out_file: (a file name)
output file for ConvertDset
argument: ``-prefix %s``, position: -1
out_type: ('niml' or 'niml_asc' or 'niml_bi' or '1D' or '1Dp' or
'1Dpt' or 'gii' or 'gii_asc' or 'gii_b64' or 'gii_b64gz')
output type
argument: ``-o_%s``, position: 0
[Optional]
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
Copy¶
Wraps the executable command 3dcopy.
Copies an image of one type to an image of the same or different type using 3dcopy command
For complete details, see the 3dcopy Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> copy3d = afni.Copy()
>>> copy3d.inputs.in_file = 'functional.nii'
>>> copy3d.cmdline
'3dcopy functional.nii functional_copy'
>>> res = copy3d.run()
>>> from copy import deepcopy
>>> copy3d_2 = deepcopy(copy3d)
>>> copy3d_2.inputs.outputtype = 'NIFTI'
>>> copy3d_2.cmdline
'3dcopy functional.nii functional_copy.nii'
>>> res = copy3d_2.run()
>>> copy3d_3 = deepcopy(copy3d)
>>> copy3d_3.inputs.outputtype = 'NIFTI_GZ'
>>> copy3d_3.cmdline
'3dcopy functional.nii functional_copy.nii.gz'
>>> res = copy3d_3.run()
>>> copy3d_4 = deepcopy(copy3d)
>>> copy3d_4.inputs.out_file = 'new_func.nii'
>>> copy3d_4.cmdline
'3dcopy functional.nii new_func.nii'
>>> res = copy3d_4.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dcopy
argument: ``%s``, position: -2
[Optional]
out_file: (a file name)
output image file name
argument: ``%s``, position: -1
verbose: (a boolean)
print progress reports
argument: ``-verb``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
Dot¶
Wraps the executable command 3dDot.
Correlation coefficient between sub-brick pairs. All datasets in in_files list will be concatenated. You can use sub-brick selectors in the file specification. Note: This program is not efficient when more than two subbricks are input. For complete details, see the 3ddot Documentation.
>>> from nipype.interfaces import afni
>>> dot = afni.Dot()
>>> dot.inputs.in_files = ['functional.nii[0]', 'structural.nii']
>>> dot.inputs.dodice = True
>>> dot.inputs.out_file = 'out.mask_ae_dice.txt'
>>> dot.cmdline
'3dDot -dodice functional.nii[0] structural.nii |& tee out.mask_ae_dice.txt'
>>> res = copy3d.run()
Inputs:
[Optional]
in_files: (a list of items which are a file name)
list of input files, possibly with subbrick selectors
argument: ``%s ...``, position: -2
out_file: (a file name)
collect output to a file
argument: `` |& tee %s``, position: -1
mask: (a file name)
Use this dataset as a mask
argument: ``-mask %s``
mrange: (a tuple of the form: (a float, a float))
Means to further restrict the voxels from 'mset' so thatonly those
mask values within this range (inclusive) willbe used.
argument: ``-mrange %s %s``
demean: (a boolean)
Remove the mean from each volume prior to computing the correlation
argument: ``-demean``
docor: (a boolean)
Return the correlation coefficient (default).
argument: ``-docor``
dodot: (a boolean)
Return the dot product (unscaled).
argument: ``-dodot``
docoef: (a boolean)
Return the least square fit coefficients {{a,b}} so that dset2 is
approximately a + b*dset1
argument: ``-docoef``
dosums: (a boolean)
Return the 6 numbers xbar=<x> ybar=<y> <(x-xbar)^2> <(y-ybar)^2>
<(x-xbar)(y-ybar)> and the correlation coefficient.
argument: ``-dosums``
dodice: (a boolean)
Return the Dice coefficient (the Sorensen-Dice index).
argument: ``-dodice``
doeta2: (a boolean)
Return eta-squared (Cohen, NeuroImage 2008).
argument: ``-doeta2``
full: (a boolean)
Compute the whole matrix. A waste of time, but handy for parsing.
argument: ``-full``
show_labels: (a boolean)
Print sub-brick labels to help identify what is being correlated.
This option is useful whenyou have more than 2 sub-bricks at input.
argument: ``-show_labels``
upper: (a boolean)
Compute upper triangular matrix
argument: ``-upper``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
Edge3¶
Wraps the executable command 3dedge3.
Does 3D Edge detection using the library 3DEdge by Gregoire Malandain (gregoire.malandain@sophia.inria.fr).
For complete details, see the 3dedge3 Documentation.
- references_ = [{‘entry’: BibTeX(‘@article{Deriche1987,’
- ‘author={R. Deriche},’ ‘title={Optimal edge detection using recursive filtering},’ ‘journal={International Journal of Computer Vision},’ ‘volume={2},’, ‘pages={167-187},’ ‘year={1987},’ ‘}’),
‘tags’: [‘method’], },
- {‘entry’: BibTeX(‘@article{MongaDericheMalandainCocquerez1991,’
- ‘author={O. Monga, R. Deriche, G. Malandain, J.P. Cocquerez},’ ‘title={Recursive filtering and edge tracking: two primary tools for 3D edge detection},’ ‘journal={Image and vision computing},’ ‘volume={9},’, ‘pages={203-214},’ ‘year={1991},’ ‘}’),
‘tags’: [‘method’], },
~
Examples¶
>>> from nipype.interfaces import afni
>>> edge3 = afni.Edge3()
>>> edge3.inputs.in_file = 'functional.nii'
>>> edge3.inputs.out_file = 'edges.nii'
>>> edge3.inputs.datum = 'byte'
>>> edge3.cmdline
'3dedge3 -input functional.nii -datum byte -prefix edges.nii'
>>> res = edge3.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dedge3
argument: ``-input %s``, position: 0
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``, position: -1
datum: ('byte' or 'short' or 'float')
specify data type for output. Valid types are 'byte', 'short' and
'float'.
argument: ``-datum %s``
fscale: (a boolean)
Force scaling of the output to the maximum integer range.
argument: ``-fscale``
mutually_exclusive: gscale, nscale, scale_floats
gscale: (a boolean)
Same as '-fscale', but also forces each output sub-brick to to get
the same scaling factor.
argument: ``-gscale``
mutually_exclusive: fscale, nscale, scale_floats
nscale: (a boolean)
Don't do any scaling on output to byte or short datasets.
argument: ``-nscale``
mutually_exclusive: fscale, gscale, scale_floats
scale_floats: (a float)
Multiply input by VAL, but only if the input datum is float. This is
needed when the input dataset has a small range, like 0 to 2.0 for
instance. With such a range, very few edges are detected due to what
I suspect to be truncation problems. Multiplying such a dataset by
10000 fixes the problem and the scaling is undone at the output.
argument: ``-scale_floats %f``
mutually_exclusive: fscale, gscale, nscale
verbose: (a boolean)
Print out some information along the way.
argument: ``-verbose``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
Eval¶
Wraps the executable command 1deval.
Evaluates an expression that may include columns of data from one or more text files.
For complete details, see the 1deval Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> eval = afni.Eval()
>>> eval.inputs.in_file_a = 'seed.1D'
>>> eval.inputs.in_file_b = 'resp.1D'
>>> eval.inputs.expr = 'a*b'
>>> eval.inputs.out1D = True
>>> eval.inputs.out_file = 'data_calc.1D'
>>> eval.cmdline
'1deval -a seed.1D -b resp.1D -expr "a*b" -1D -prefix data_calc.1D'
>>> res = eval.run()
Inputs:
[Mandatory]
in_file_a: (an existing file name)
input file to 1deval
argument: ``-a %s``, position: 0
expr: (a unicode string)
expr
argument: ``-expr "%s"``, position: 3
[Optional]
in_file_b: (an existing file name)
operand file to 1deval
argument: ``-b %s``, position: 1
in_file_c: (an existing file name)
operand file to 1deval
argument: ``-c %s``, position: 2
out_file: (a file name)
output image file name
argument: ``-prefix %s``
out1D: (a boolean)
output in 1D
argument: ``-1D``
start_idx: (an integer (int or long))
start index for in_file_a
requires: stop_idx
stop_idx: (an integer (int or long))
stop index for in_file_a
requires: start_idx
single_idx: (an integer (int or long))
volume index for in_file_a
other: (a file name)
other options
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
FWHMx¶
Wraps the executable command 3dFWHMx.
Unlike the older 3dFWHM, this program computes FWHMs for all sub-bricks in the input dataset, each one separately. The output for each one is written to the file specified by ‘-out’. The mean (arithmetic or geometric) of all the FWHMs along each axis is written to stdout. (A non-positive output value indicates something bad happened; e.g., FWHM in z is meaningless for a 2D dataset; the estimation method computed incoherent intermediate results.)
For complete details, see the 3dFWHMx Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> fwhm = afni.FWHMx()
>>> fwhm.inputs.in_file = 'functional.nii'
>>> fwhm.cmdline
'3dFWHMx -input functional.nii -out functional_subbricks.out > functional_fwhmx.out'
>>> res = fwhm.run()
(Classic) METHOD:
- Calculate ratio of variance of first differences to data variance.
- Should be the same as 3dFWHM for a 1-brick dataset. (But the output format is simpler to use in a script.)
Note
IMPORTANT NOTE [AFNI > 16]
A completely new method for estimating and using noise smoothness values is now available in 3dFWHMx and 3dClustSim. This method is implemented in the ‘-acf’ options to both programs. ‘ACF’ stands for (spatial) AutoCorrelation Function, and it is estimated by calculating moments of differences out to a larger radius than before.
Notably, real FMRI data does not actually have a Gaussian-shaped ACF, so the estimated ACF is then fit (in 3dFWHMx) to a mixed model (Gaussian plus mono-exponential) of the form
where  is the radius, and
is the radius, and 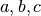 are the fitted parameters.
The apparent FWHM from this model is usually somewhat larger in real data
than the FWHM estimated from just the nearest-neighbor differences used
in the ‘classic’ analysis.
are the fitted parameters.
The apparent FWHM from this model is usually somewhat larger in real data
than the FWHM estimated from just the nearest-neighbor differences used
in the ‘classic’ analysis.
The longer tails provided by the mono-exponential are also significant. 3dClustSim has also been modified to use the ACF model given above to generate noise random fields.
Note
TL;DR or summary
The take-awaymessage is that the ‘classic’ 3dFWHMx and 3dClustSim analysis, using a pure Gaussian ACF, is not very correct for FMRI data – I cannot speak for PET or MEG data.
Warning
Do NOT use 3dFWHMx on the statistical results (e.g., ‘-bucket’) from 3dDeconvolve or 3dREMLfit!!! The function of 3dFWHMx is to estimate the smoothness of the time series NOISE, not of the statistics. This proscription is especially true if you plan to use 3dClustSim next!!
Note
Recommendations
- For FMRI statistical purposes, you DO NOT want the FWHM to reflect the spatial structure of the underlying anatomy. Rather, you want the FWHM to reflect the spatial structure of the noise. This means that the input dataset should not have anatomical (spatial) structure.
- One good form of input is the output of ‘3dDeconvolve -errts’, which is the dataset of residuals left over after the GLM fitted signal model is subtracted out from each voxel’s time series.
- If you don’t want to go to that much trouble, use ‘-detrend’ to approximately subtract out the anatomical spatial structure, OR use the output of 3dDetrend for the same purpose.
- If you do not use ‘-detrend’, the program attempts to find non-zero spatial structure in the input, and will print a warning message if it is detected.
Note
Notes on -demend
- I recommend this option, and it is not the default only for historical compatibility reasons. It may become the default someday.
- It is already the default in program 3dBlurToFWHM. This is the same detrending as done in 3dDespike; using 2*q+3 basis functions for q > 0.
- If you don’t use ‘-detrend’, the program now [Aug 2010] checks if a large number of voxels are have significant nonzero means. If so, the program will print a warning message suggesting the use of ‘-detrend’, since inherent spatial structure in the image will bias the estimation of the FWHM of the image time series NOISE (which is usually the point of using 3dFWHMx).
Inputs:
[Mandatory]
in_file: (an existing file name)
input dataset
argument: ``-input %s``
[Optional]
out_file: (a file name)
output file
argument: ``> %s``, position: -1
out_subbricks: (a file name)
output file listing the subbricks FWHM
argument: ``-out %s``
mask: (an existing file name)
use only voxels that are nonzero in mask
argument: ``-mask %s``
automask: (a boolean, nipype default value: False)
compute a mask from THIS dataset, a la 3dAutomask
argument: ``-automask``
detrend: (a boolean or an integer (int or long), nipype default
value: False)
instead of demed (0th order detrending), detrend to the specified
order. If order is not given, the program picks q=NT/30. -detrend
disables -demed, and includes -unif.
argument: ``-detrend``
mutually_exclusive: demed
demed: (a boolean)
If the input dataset has more than one sub-brick (e.g., has a time
axis), then subtract the median of each voxel's time series before
processing FWHM. This will tend to remove intrinsic spatial
structure and leave behind the noise.
argument: ``-demed``
mutually_exclusive: detrend
unif: (a boolean)
If the input dataset has more than one sub-brick, then normalize
each voxel's time series to have the same MAD before processing
FWHM.
argument: ``-unif``
out_detrend: (a file name)
Save the detrended file into a dataset
argument: ``-detprefix %s``
geom: (a boolean)
if in_file has more than one sub-brick, compute the final estimate
as the geometric mean of the individual sub-brick FWHM estimates
argument: ``-geom``
mutually_exclusive: arith
arith: (a boolean)
if in_file has more than one sub-brick, compute the final estimate
as the arithmetic mean of the individual sub-brick FWHM estimates
argument: ``-arith``
mutually_exclusive: geom
combine: (a boolean)
combine the final measurements along each axis
argument: ``-combine``
compat: (a boolean)
be compatible with the older 3dFWHM
argument: ``-compat``
acf: (a boolean or a file name or a tuple of the form: (an existing
file name, a float), nipype default value: False)
computes the spatial autocorrelation
argument: ``-acf``
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
out_subbricks: (an existing file name)
output file (subbricks)
out_detrend: (a file name)
output file, detrended
fwhm: (a tuple of the form: (a float, a float, a float) or a tuple of
the form: (a float, a float, a float, a float))
FWHM along each axis
acf_param: (a tuple of the form: (a float, a float, a float) or a
tuple of the form: (a float, a float, a float, a float))
fitted ACF model parameters
out_acf: (an existing file name)
output acf file
References:¶
None
GCOR¶
Wraps the executable command @compute_gcor.
Computes the average correlation between every voxel and ever other voxel, over any give mask.
For complete details, see the @compute_gcor Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> gcor = afni.GCOR()
>>> gcor.inputs.in_file = 'structural.nii'
>>> gcor.inputs.nfirst = 4
>>> gcor.cmdline
'@compute_gcor -nfirst 4 -input structural.nii'
>>> res = gcor.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input dataset to compute the GCOR over
argument: ``-input %s``, position: -1
[Optional]
mask: (an existing file name)
mask dataset, for restricting the computation
argument: ``-mask %s``
nfirst: (an integer (int or long))
specify number of initial TRs to ignore
argument: ``-nfirst %d``
no_demean: (a boolean)
do not (need to) demean as first step
argument: ``-no_demean``
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out: (a float)
global correlation value
LocalBistat¶
Wraps the executable command 3dLocalBistat.
3dLocalBistat - computes statistics between 2 datasets, at each voxel, based on a local neighborhood of that voxel.
For complete details, see the 3dLocalBistat Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> bistat = afni.LocalBistat()
>>> bistat.inputs.in_file1 = 'functional.nii'
>>> bistat.inputs.in_file2 = 'structural.nii'
>>> bistat.inputs.neighborhood = ('SPHERE', 1.2)
>>> bistat.inputs.stat = 'pearson'
>>> bistat.inputs.outputtype = 'NIFTI'
>>> bistat.cmdline
"3dLocalBistat -prefix functional_bistat.nii -nbhd 'SPHERE(1.2)' -stat pearson functional.nii structural.nii"
>>> res = automask.run()
Inputs:
[Mandatory]
in_file1: (an existing file name)
Filename of the first image
argument: ``%s``, position: -2
in_file2: (an existing file name)
Filename of the second image
argument: ``%s``, position: -1
neighborhood: (a tuple of the form: ('SPHERE' or 'RHDD' or 'TOHD', a
float) or a tuple of the form: ('RECT', a tuple of the form: (a
float, a float, a float)))
The region around each voxel that will be extracted for the
statistics calculation. Possible regions are: 'SPHERE', 'RHDD'
(rhombic dodecahedron), 'TOHD' (truncated octahedron) with a given
radius in mm or 'RECT' (rectangular block) with dimensions to
specify in mm.
argument: ``-nbhd '%s(%s)'``
stat: (a list of items which are 'pearson' or 'spearman' or
'quadrant' or 'mutinfo' or 'normuti' or 'jointent' or 'hellinger'
or 'crU' or 'crM' or 'crA' or 'L2slope' or 'L1slope' or 'num' or
'ALL')
statistics to compute. Possible names are : * pearson = Pearson
correlation coefficient * spearman = Spearman correlation
coefficient * quadrant = Quadrant correlation coefficient * mutinfo
= Mutual Information * normuti = Normalized Mutual Information *
jointent = Joint entropy * hellinger= Hellinger metric * crU =
Correlation ratio (Unsymmetric) * crM = Correlation ratio
(symmetrized by Multiplication) * crA = Correlation ratio
(symmetrized by Addition) * L2slope = slope of least-squares (L2)
linear regression of the data from dataset1 vs. the dataset2 (i.e.,
d2 = a + b*d1 ==> this is 'b') * L1slope = slope of least-absolute-
sum (L1) linear regression of the data from dataset1 vs. the
dataset2 * num = number of the values in the region: with the use of
-mask or -automask, the size of the region around any given voxel
will vary; this option lets you map that size. * ALL = all of the
above, in that orderMore than one option can be used.
argument: ``-stat %s...``
[Optional]
mask_file: (a file name)
mask image file name. Voxels NOT in the mask will not be used in the
neighborhood of any voxel. Also, a voxel NOT in the mask will have
its statistic(s) computed as zero (0).
argument: ``-mask %s``
automask: (a boolean)
Compute the mask as in program 3dAutomask.
argument: ``-automask``
mutually_exclusive: weight_file
weight_file: (a file name)
File name of an image to use as a weight. Only applies to 'pearson'
statistics.
argument: ``-weight %s``
mutually_exclusive: automask
out_file: (a file name)
Output dataset.
argument: ``-prefix %s``, position: 0
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
Localstat¶
Wraps the executable command 3dLocalstat.
3dLocalstat - computes statistics at each voxel, based on a local neighborhood of that voxel.
For complete details, see the 3dLocalstat Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> localstat = afni.Localstat()
>>> localstat.inputs.in_file = 'functional.nii'
>>> localstat.inputs.mask_file = 'skeleton_mask.nii.gz'
>>> localstat.inputs.neighborhood = ('SPHERE', 45)
>>> localstat.inputs.stat = 'mean'
>>> localstat.inputs.nonmask = True
>>> localstat.inputs.outputtype = 'NIFTI_GZ'
>>> localstat.cmdline
"3dLocalstat -prefix functional_localstat.nii -mask skeleton_mask.nii.gz -nbhd 'SPHERE(45.0)' -use_nonmask -stat mean functional.nii"
>>> res = localstat.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input dataset
argument: ``%s``, position: -1
neighborhood: (a tuple of the form: ('SPHERE' or 'RHDD' or 'TOHD', a
float) or a tuple of the form: ('RECT', a tuple of the form: (a
float, a float, a float)))
The region around each voxel that will be extracted for the
statistics calculation. Possible regions are: 'SPHERE', 'RHDD'
(rhombic dodecahedron), 'TOHD' (truncated octahedron) with a given
radius in mm or 'RECT' (rectangular block) with dimensions to
specify in mm.
argument: ``-nbhd '%s(%s)'``
stat: (a list of items which are 'mean' or 'stdev' or 'var' or 'cvar'
or 'median' or 'MAD' or 'min' or 'max' or 'absmax' or 'num' or
'sum' or 'FWHM' or 'FWHMbar' or 'rank' or 'frank' or 'P2skew' or
'ALL' or 'mMP2s' or 'mmMP2s' or a tuple of the form: ('perc', a
tuple of the form: (a float, a float, a float)))
statistics to compute. Possible names are :
* mean = average of the values
* stdev = standard deviation
* var = variance (stdev*stdev)
* cvar = coefficient of variation = stdev/fabs(mean)
* median = median of the values
* MAD = median absolute deviation
* min = minimum
* max = maximum
* absmax = maximum of the absolute values
* num = number of the values in the region:
with the use of -mask or -automask, the size of the region around
any given voxel will vary; this option lets you map that size. It
may be useful if you plan to compute a t-statistic (say) from the
mean and stdev outputs.
* sum = sum of the values in the region
* FWHM = compute (like 3dFWHM) image smoothness inside each voxel's
neighborhood. Results are in 3 sub-bricks: FWHMx, FHWMy, and FWHMz.
Places where an output is -1 are locations where the FWHM value
could not be computed (e.g., outside the mask).
* FWHMbar= Compute just the average of the 3 FWHM values (normally
would NOT do this with FWHM also).
* perc:P0:P1:Pstep =
Compute percentiles between P0 and P1 with a step of Pstep.
Default P1 is equal to P0 and default P2 = 1
* rank = rank of the voxel's intensity
* frank = rank / number of voxels in neighborhood
* P2skew = Pearson's second skewness coefficient 3 * (mean -
median) / stdev
* ALL = all of the above, in that order (except for FWHMbar and
perc).
* mMP2s = Exactly the same output as: median, MAD, P2skew, but a
little faster
* mmMP2s = Exactly the same output as: mean, median, MAD, P2skew
More than one option can be used.
argument: ``-stat %s...``
[Optional]
mask_file: (a file name)
Mask image file name. Voxels NOT in the mask will not be used in the
neighborhood of any voxel. Also, a voxel NOT in the mask will have
its statistic(s) computed as zero (0) unless the parameter 'nonmask'
is set to true.
argument: ``-mask %s``
automask: (a boolean)
Compute the mask as in program 3dAutomask.
argument: ``-automask``
nonmask: (a boolean)
Voxels not in the mask WILL have their local statistics computed
from all voxels in their neighborhood that ARE in the mask.
* For instance, this option can be used to compute the average
local white matter time series, even at non-WM voxels.
argument: ``-use_nonmask``
reduce_grid: (a float or a tuple of the form: (a float, a float, a
float))
Compute output on a grid that is reduced by the specified factors.
If a single value is passed, output is resampled to the specified
isotropic grid. Otherwise, the 3 inputs describe the reduction in
the X, Y, and Z directions. This option speeds up computations at
the expense of resolution. It should only be used when the nbhd is
quite large with respect to the input's resolution, and the
resultant stats are expected to be smooth.
argument: ``-reduce_grid %s``
mutually_exclusive: reduce_restore_grid, reduce_max_vox
reduce_restore_grid: (a float or a tuple of the form: (a float, a
float, a float))
Like reduce_grid, but also resample output back to inputgrid.
argument: ``-reduce_restore_grid %s``
mutually_exclusive: reduce_max_vox, reduce_grid
reduce_max_vox: (a float)
Like reduce_restore_grid, but automatically set Rx Ry Rz sothat the
computation grid is at a resolution of nbhd/MAX_VOXvoxels.
argument: ``-reduce_max_vox %s``
mutually_exclusive: reduce_restore_grid, reduce_grid
grid_rmode: ('NN' or 'Li' or 'Cu' or 'Bk')
Interpolant to use when resampling the output with
thereduce_restore_grid option. The resampling method string RESAM
should come from the set {'NN', 'Li', 'Cu', 'Bk'}. These stand for
'Nearest Neighbor', 'Linear', 'Cubic', and 'Blocky' interpolation,
respectively.
argument: ``-grid_rmode %s``
requires: reduce_restore_grid
quiet: (a boolean)
Stop the highly informative progress reports.
argument: ``-quiet``
overwrite: (a boolean)
overwrite output file if it already exists
argument: ``-overwrite``
out_file: (a file name)
Output dataset.
argument: ``-prefix %s``, position: 0
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
MaskTool¶
Wraps the executable command 3dmask_tool.
3dmask_tool - for combining/dilating/eroding/filling masks
For complete details, see the 3dmask_tool Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> masktool = afni.MaskTool()
>>> masktool.inputs.in_file = 'functional.nii'
>>> masktool.inputs.outputtype = 'NIFTI'
>>> masktool.cmdline
'3dmask_tool -prefix functional_mask.nii -input functional.nii'
>>> res = automask.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file or files to 3dmask_tool
argument: ``-input %s``, position: -1
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``
count: (a boolean)
Instead of created a binary 0/1 mask dataset, create one with counts
of voxel overlap, i.e., each voxel will contain the number of masks
that it is set in.
argument: ``-count``, position: 2
datum: ('byte' or 'short' or 'float')
specify data type for output. Valid types are 'byte', 'short' and
'float'.
argument: ``-datum %s``
dilate_inputs: (a unicode string)
Use this option to dilate and/or erode datasets as they are read.
ex. '5 -5' to dilate and erode 5 times
argument: ``-dilate_inputs %s``
dilate_results: (a unicode string)
dilate and/or erode combined mask at the given levels.
argument: ``-dilate_results %s``
frac: (a float)
When combining masks (across datasets and sub-bricks), use this
option to restrict the result to a certain fraction of the set of
volumes
argument: ``-frac %s``
inter: (a boolean)
intersection, this means -frac 1.0
argument: ``-inter``
union: (a boolean)
union, this means -frac 0
argument: ``-union``
fill_holes: (a boolean)
This option can be used to fill holes in the resulting mask, i.e.
after all other processing has been done.
argument: ``-fill_holes``
fill_dirs: (a unicode string)
fill holes only in the given directions. This option is for use with
-fill holes. should be a single string that specifies 1-3 of the
axes using {x,y,z} labels (i.e. dataset axis order), or using the
labels in {R,L,A,P,I,S}.
argument: ``-fill_dirs %s``
requires: fill_holes
verbose: (an integer (int or long))
specify verbosity level, for 0 to 3
argument: ``-verb %s``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
mask file
References:¶
None None
Merge¶
Wraps the executable command 3dmerge.
Merge or edit volumes using AFNI 3dmerge command
For complete details, see the 3dmerge Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> merge = afni.Merge()
>>> merge.inputs.in_files = ['functional.nii', 'functional2.nii']
>>> merge.inputs.blurfwhm = 4
>>> merge.inputs.doall = True
>>> merge.inputs.out_file = 'e7.nii'
>>> merge.cmdline
'3dmerge -1blur_fwhm 4 -doall -prefix e7.nii functional.nii functional2.nii'
>>> res = merge.run()
Inputs:
[Mandatory]
in_files: (a list of items which are an existing file name)
argument: ``%s``, position: -1
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``
doall: (a boolean)
apply options to all sub-bricks in dataset
argument: ``-doall``
blurfwhm: (an integer (int or long))
FWHM blur value (mm)
argument: ``-1blur_fwhm %d``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
Notes¶
Wraps the executable command 3dNotes.
A program to add, delete, and show notes for AFNI datasets.
For complete details, see the 3dNotes Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> notes = afni.Notes()
>>> notes.inputs.in_file = 'functional.HEAD'
>>> notes.inputs.add = 'This note is added.'
>>> notes.inputs.add_history = 'This note is added to history.'
>>> notes.cmdline
'3dNotes -a "This note is added." -h "This note is added to history." functional.HEAD'
>>> res = notes.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dNotes
argument: ``%s``, position: -1
[Optional]
add: (a unicode string)
note to add
argument: ``-a "%s"``
add_history: (a unicode string)
note to add to history
argument: ``-h "%s"``
mutually_exclusive: rep_history
rep_history: (a unicode string)
note with which to replace history
argument: ``-HH "%s"``
mutually_exclusive: add_history
delete: (an integer (int or long))
delete note number num
argument: ``-d %d``
ses: (a boolean)
print to stdout the expanded notes
argument: ``-ses``
out_file: (a file name)
output image file name
argument: ``%s``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
NwarpAdjust¶
Wraps the executable command 3dNwarpAdjust.
This program takes as input a bunch of 3D warps, averages them, and computes the inverse of this average warp. It then composes each input warp with this inverse average to ‘adjust’ the set of warps. Optionally, it can also read in a set of 1-brick datasets corresponding to the input warps, and warp each of them, and average those.
For complete details, see the 3dNwarpAdjust Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> adjust = afni.NwarpAdjust()
>>> adjust.inputs.warps = ['func2anat_InverseWarp.nii.gz', 'func2anat_InverseWarp.nii.gz', 'func2anat_InverseWarp.nii.gz', 'func2anat_InverseWarp.nii.gz', 'func2anat_InverseWarp.nii.gz']
>>> adjust.cmdline
'3dNwarpAdjust -nwarp func2anat_InverseWarp.nii.gz func2anat_InverseWarp.nii.gz func2anat_InverseWarp.nii.gz func2anat_InverseWarp.nii.gz func2anat_InverseWarp.nii.gz'
>>> res = adjust.run()
Inputs:
[Mandatory]
warps: (a list of at least 5 items which are an existing file name)
List of input 3D warp datasets
argument: ``-nwarp %s``
[Optional]
in_files: (a list of at least 5 items which are an existing file
name)
List of input 3D datasets to be warped by the adjusted warp
datasets. There must be exactly as many of these datasets as there
are input warps.
argument: ``-source %s``
out_file: (a file name)
Output mean dataset, only needed if in_files are also given. The
output dataset will be on the common grid shared by the source
datasets.
argument: ``-prefix %s``
requires: in_files
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
NwarpApply¶
Wraps the executable command 3dNwarpApply.
Program to apply a nonlinear 3D warp saved from 3dQwarp (or 3dNwarpCat, etc.) to a 3D dataset, to produce a warped version of the source dataset.
For complete details, see the 3dNwarpApply Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> nwarp = afni.NwarpApply()
>>> nwarp.inputs.in_file = 'Fred+orig'
>>> nwarp.inputs.master = 'NWARP'
>>> nwarp.inputs.warp = "'Fred_WARP+tlrc Fred.Xaff12.1D'"
>>> nwarp.cmdline
"3dNwarpApply -source Fred+orig -interp wsinc5 -master NWARP -prefix Fred+orig_Nwarp -nwarp 'Fred_WARP+tlrc Fred.Xaff12.1D'"
>>> res = nwarp.run()
Inputs:
[Mandatory]
in_file: (an existing file name or a list of items which are an
existing file name)
the name of the dataset to be warped can be multiple datasets
argument: ``-source %s``
warp: (a string)
the name of the warp dataset. multiple warps can be concatenated
(make sure they exist)
argument: ``-nwarp %s``
[Optional]
inv_warp: (a boolean)
After the warp specified in '-nwarp' is computed, invert it
argument: ``-iwarp``
master: (a file name)
the name of the master dataset, which defines the output grid
argument: ``-master %s``
interp: ('wsinc5' or 'NN' or 'nearestneighbour' or 'nearestneighbor'
or 'linear' or 'trilinear' or 'cubic' or 'tricubic' or 'quintic'
or 'triquintic', nipype default value: wsinc5)
defines interpolation method to use during warp
argument: ``-interp %s``
ainterp: ('NN' or 'nearestneighbour' or 'nearestneighbor' or 'linear'
or 'trilinear' or 'cubic' or 'tricubic' or 'quintic' or
'triquintic' or 'wsinc5')
specify a different interpolation method than might be used for the
warp
argument: ``-ainterp %s``
out_file: (a file name)
output image file name
argument: ``-prefix %s``
short: (a boolean)
Write output dataset using 16-bit short integers, rather than the
usual 32-bit floats.
argument: ``-short``
quiet: (a boolean)
don't be verbose :(
argument: ``-quiet``
mutually_exclusive: verb
verb: (a boolean)
be extra verbose :)
argument: ``-verb``
mutually_exclusive: quiet
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
NwarpCat¶
Wraps the executable command 3dNwarpCat.
Catenates (composes) 3D warps defined on a grid, OR via a matrix.
Note
All transformations are from DICOM xyz (in mm) to DICOM xyz.
Matrix warps are in files that end in ‘.1D’ or in ‘.txt’. A matrix warp file should have 12 numbers in it, as output (for example), by ‘3dAllineate -1Dmatrix_save’.
Nonlinear warps are in dataset files (AFNI .HEAD/.BRIK or NIfTI .nii) with 3 sub-bricks giving the DICOM order xyz grid displacements in mm.
If all the input warps are matrices, then the output is a matrix and will be written to the file ‘prefix.aff12.1D’. Unless the prefix already contains the string ‘.1D’, in which case the filename is just the prefix.
If ‘prefix’ is just ‘stdout’, then the output matrix is written to standard output. In any of these cases, the output format is 12 numbers in one row.
If any of the input warps are datasets, they must all be defined on the same 3D grid! And of course, then the output will be a dataset on the same grid. However, you can expand the grid using the ‘-expad’ option.
The order of operations in the final (output) warp is, for the case of 3 input warps:
OUTPUT(x) = warp3( warp2( warp1(x) ) )
That is, warp1 is applied first, then warp2, et cetera. The 3D x coordinates are taken from each grid location in the first dataset defined on a grid.
For complete details, see the 3dNwarpCat Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> nwarpcat = afni.NwarpCat()
>>> nwarpcat.inputs.in_files = ['Q25_warp+tlrc.HEAD', ('IDENT', 'structural.nii')]
>>> nwarpcat.inputs.out_file = 'Fred_total_WARP'
>>> nwarpcat.cmdline
"3dNwarpCat -interp wsinc5 -prefix Fred_total_WARP Q25_warp+tlrc.HEAD 'IDENT(structural.nii)'"
>>> res = nwarpcat.run()
Inputs:
[Mandatory]
in_files: (a list of items which are a file name or a tuple of the
form: ('IDENT' or 'INV' or 'SQRT' or 'SQRTINV', a file name))
list of tuples of 3D warps and associated functions
argument: ``%s``, position: -1
[Optional]
space: (a string)
string to attach to the output dataset as its atlas space marker.
argument: ``-space %s``
inv_warp: (a boolean)
invert the final warp before output
argument: ``-iwarp``
interp: ('wsinc5' or 'linear' or 'quintic', nipype default value:
wsinc5)
specify a different interpolation method than might be used for the
warp
argument: ``-interp %s``
expad: (an integer (int or long))
Pad the nonlinear warps by the given number of voxels voxels in all
directions. The warp displacements are extended by linear
extrapolation from the faces of the input grid..
argument: ``-expad %d``
out_file: (a file name)
output image file name
argument: ``-prefix %s``
verb: (a boolean)
be verbose
argument: ``-verb``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
OneDToolPy¶
Wraps the executable command 1d_tool.py.
This program is meant to read/manipulate/write/diagnose 1D datasets. Input can be specified using AFNI sub-brick[]/time{} selectors.
>>> from nipype.interfaces import afni
>>> odt = afni.OneDToolPy()
>>> odt.inputs.in_file = 'f1.1D'
>>> odt.inputs.set_nruns = 3
>>> odt.inputs.demean = True
>>> odt.inputs.out_file = 'motion_dmean.1D'
>>> odt.cmdline
'python2 ...1d_tool.py -demean -infile f1.1D -write motion_dmean.1D -set_nruns 3'
>>> res = odt.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to OneDTool
argument: ``-infile %s``
[Optional]
set_nruns: (an integer (int or long))
treat the input data as if it has nruns
argument: ``-set_nruns %d``
derivative: (a boolean)
take the temporal derivative of each vector (done as first backward
difference)
argument: ``-derivative``
demean: (a boolean)
demean each run (new mean of each run = 0.0)
argument: ``-demean``
out_file: (a file name)
write the current 1D data to FILE
argument: ``-write %s``
mutually_exclusive: show_cormat_warnings
show_censor_count: (a boolean)
display the total number of censored TRs Note : if input is a valid
xmat.1D dataset, then the count will come from the header. Otherwise
the input is assumed to be a binary censorfile, and zeros are simply
counted.
argument: ``-show_censor_count``
censor_motion: (a tuple of the form: (a float, a file name))
Tuple of motion limit and outfile prefix. need to also set set_nruns
-r set_run_lengths
argument: ``-censor_motion %f %s``
censor_prev_TR: (a boolean)
for each censored TR, also censor previous
argument: ``-censor_prev_TR``
show_trs_uncensored: ('comma' or 'space' or 'encoded' or 'verbose')
display a list of TRs which were not censored in the specified style
argument: ``-show_trs_uncensored %s``
show_cormat_warnings: (a file name)
Write cormat warnings to a file
argument: ``-show_cormat_warnings |& tee %s``, position: -1
mutually_exclusive: out_file
show_indices_interest: (a boolean)
display column indices for regs of interest
argument: ``-show_indices_interest``
show_trs_run: (an integer (int or long))
restrict -show_trs_[un]censored to the given 1-based run
argument: ``-show_trs_run %d``
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
py27_path: (an existing file name or 'python2', nipype default value:
python2)
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (a file name)
output of 1D_tool.py
References:¶
None None
ReHo¶
Wraps the executable command 3dReHo.
Compute regional homogenity for a given neighbourhood.l, based on a local neighborhood of that voxel.
For complete details, see the 3dReHo Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> reho = afni.ReHo()
>>> reho.inputs.in_file = 'functional.nii'
>>> reho.inputs.out_file = 'reho.nii.gz'
>>> reho.inputs.neighborhood = 'vertices'
>>> reho.cmdline
'3dReHo -prefix reho.nii.gz -inset functional.nii -nneigh 27'
>>> res = reho.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input dataset
argument: ``-inset %s``, position: 1
[Optional]
out_file: (a file name)
Output dataset.
argument: ``-prefix %s``, position: 0
chi_sq: (a boolean)
Output the Friedman chi-squared value in addition to the Kendall's
W. This option is currently compatible only with the AFNI
(BRIK/HEAD) output type; the chi-squared value will be the second
sub-brick of the output dataset.
argument: ``-chi_sq``
mask_file: (a file name)
Mask within which ReHo should be calculated voxelwise
argument: ``-mask %s``
neighborhood: ('faces' or 'edges' or 'vertices')
voxels in neighborhood. can be: * faces (for voxel and 6 facewise
neighbors, only),
* edges (for voxel and 18 face- and edge-wise neighbors),
* vertices (for voxel and 26 face-, edge-, and node-wise neighbors).
argument: ``-nneigh %s``
mutually_exclusive: sphere, ellipsoid
sphere: (a float)
for additional voxelwise neighborhood control, the radius R of a
desired neighborhood can be put in; R is a floating point number,
and must be >1. Examples of the numbers of voxels in a given radius
are as follows (you can roughly approximate with the ol'
4*PI*(R^3)/3 thing):
R=2.0 -> V=33,
R=2.3 -> V=57,
R=2.9 -> V=93,
R=3.1 -> V=123,
R=3.9 -> V=251,
R=4.5 -> V=389,
R=6.1 -> V=949,
but you can choose most any value.
argument: ``-neigh_RAD %s``
mutually_exclusive: neighborhood, ellipsoid
ellipsoid: (a tuple of the form: (a float, a float, a float))
Tuple indicating the x, y, and z radius of an ellipsoid defining the
neighbourhood of each voxel.
The 'hood is then made according to the following relation:(i/A)^2 +
(j/B)^2 + (k/C)^2 <=1.
which will have approx. V=4*PI*A*B*C/3. The impetus for this freedom
was for use with data having anisotropic voxel edge lengths.
argument: ``-neigh_X %s -neigh_Y %s -neigh_Z %s``
mutually_exclusive: sphere, neighborhood
label_set: (an existing file name)
a set of ROIs, each labelled with distinct integers. ReHo will then
be calculated per ROI.
argument: ``-in_rois %s``
overwrite: (a boolean)
overwrite output file if it already exists
argument: ``-overwrite``
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
Voxelwise regional homogeneity map
out_vals: (a file name)
Table of labelwise regional homogenity values
Refit¶
Wraps the executable command 3drefit.
Changes some of the information inside a 3D dataset’s header
For complete details, see the 3drefit Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> refit = afni.Refit()
>>> refit.inputs.in_file = 'structural.nii'
>>> refit.inputs.deoblique = True
>>> refit.cmdline
'3drefit -deoblique structural.nii'
>>> res = refit.run()
>>> refit_2 = afni.Refit()
>>> refit_2.inputs.in_file = 'structural.nii'
>>> refit_2.inputs.atrfloat = ("IJK_TO_DICOM_REAL", "'1 0.2 0 0 -0.2 1 0 0 0 0 1 0'")
>>> refit_2.cmdline
"3drefit -atrfloat IJK_TO_DICOM_REAL '1 0.2 0 0 -0.2 1 0 0 0 0 1 0' structural.nii"
>>> res = refit_2.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3drefit
argument: ``%s``, position: -1
[Optional]
deoblique: (a boolean)
replace current transformation matrix with cardinal matrix
argument: ``-deoblique``
xorigin: (a unicode string)
x distance for edge voxel offset
argument: ``-xorigin %s``
yorigin: (a unicode string)
y distance for edge voxel offset
argument: ``-yorigin %s``
zorigin: (a unicode string)
z distance for edge voxel offset
argument: ``-zorigin %s``
duporigin_file: (an existing file name)
Copies the xorigin, yorigin, and zorigin values from the header of
the given dataset
argument: ``-duporigin %s``
xdel: (a float)
new x voxel dimension in mm
argument: ``-xdel %f``
ydel: (a float)
new y voxel dimension in mm
argument: ``-ydel %f``
zdel: (a float)
new z voxel dimension in mm
argument: ``-zdel %f``
xyzscale: (a float)
Scale the size of the dataset voxels by the given factor
argument: ``-xyzscale %f``
space: ('TLRC' or 'MNI' or 'ORIG')
Associates the dataset with a specific template type, e.g. TLRC,
MNI, ORIG
argument: ``-space %s``
atrcopy: (a tuple of the form: (a file name, a unicode string))
Copy AFNI header attribute from the given file into the header of
the dataset(s) being modified. For more information on AFNI header
attributes, see documentation file README.attributes. More than one
'-atrcopy' option can be used. For AFNI advanced users only. Do NOT
use -atrcopy or -atrstring with other modification options. See also
-copyaux.
argument: ``-atrcopy %s %s``
atrstring: (a tuple of the form: (a unicode string, a unicode
string))
Copy the last given string into the dataset(s) being modified,
giving it the attribute name given by the last string.To be safe,
the last string should be in quotes.
argument: ``-atrstring %s %s``
atrfloat: (a tuple of the form: (a unicode string, a unicode string))
Create or modify floating point attributes. The input values may be
specified as a single string in quotes or as a 1D filename or
string, example '1 0.2 0 0 -0.2 1 0 0 0 0 1 0' or flipZ.1D or
'1D:1,0.2,2@0,-0.2,1,2@0,2@0,1,0'
argument: ``-atrfloat %s %s``
atrint: (a tuple of the form: (a unicode string, a unicode string))
Create or modify integer attributes. The input values may be
specified as a single string in quotes or as a 1D filename or
string, example '1 0 0 0 0 1 0 0 0 0 1 0' or flipZ.1D or
'1D:1,0,2@0,-0,1,2@0,2@0,1,0'
argument: ``-atrint %s %s``
saveatr: (a boolean)
(default) Copy the attributes that are known to AFNI into the
dset->dblk structure thereby forcing changes to known attributes to
be present in the output. This option only makes sense with
-atrcopy.
argument: ``-saveatr``
nosaveatr: (a boolean)
Opposite of -saveatr
argument: ``-nosaveatr``
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
Resample¶
Wraps the executable command 3dresample.
Resample or reorient an image using AFNI 3dresample command
For complete details, see the 3dresample Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> resample = afni.Resample()
>>> resample.inputs.in_file = 'functional.nii'
>>> resample.inputs.orientation= 'RPI'
>>> resample.inputs.outputtype = 'NIFTI'
>>> resample.cmdline
'3dresample -orient RPI -prefix functional_resample.nii -inset functional.nii'
>>> res = resample.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dresample
argument: ``-inset %s``, position: -1
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``
orientation: (a unicode string)
new orientation code
argument: ``-orient %s``
resample_mode: ('NN' or 'Li' or 'Cu' or 'Bk')
resampling method from set {"NN", "Li", "Cu", "Bk"}. These are for
"Nearest Neighbor", "Linear", "Cubic" and "Blocky"interpolation,
respectively. Default is NN.
argument: ``-rmode %s``
voxel_size: (a tuple of the form: (a float, a float, a float))
resample to new dx, dy and dz
argument: ``-dxyz %f %f %f``
master: (a file name)
align dataset grid to a reference file
argument: ``-master %s``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
TCat¶
Wraps the executable command 3dTcat.
Concatenate sub-bricks from input datasets into one big 3D+time dataset.
TODO Replace InputMultiPath in_files with Traits.List, if possible. Current version adds extra whitespace.
For complete details, see the 3dTcat Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> tcat = afni.TCat()
>>> tcat.inputs.in_files = ['functional.nii', 'functional2.nii']
>>> tcat.inputs.out_file= 'functional_tcat.nii'
>>> tcat.inputs.rlt = '+'
>>> tcat.cmdline
'3dTcat -rlt+ -prefix functional_tcat.nii functional.nii functional2.nii'
>>> res = tcat.run()
Inputs:
[Mandatory]
in_files: (a list of items which are an existing file name)
input file to 3dTcat
argument: `` %s``, position: -1
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``
rlt: ('' or '+' or '++')
Remove linear trends in each voxel time series loaded from each
input dataset, SEPARATELY. Option -rlt removes the least squares fit
of 'a+b*t' to each voxel time series. Option -rlt+ adds dataset mean
back in. Option -rlt++ adds overall mean of all dataset timeseries
back in.
argument: ``-rlt%s``, position: 1
verbose: (a boolean)
Print out some verbose output as the program
argument: ``-verb``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
TCatSubBrick¶
Wraps the executable command 3dTcat.
Hopefully a temporary function to allow sub-brick selection until afni file managment is improved.
For complete details, see the 3dTcat Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> tcsb = afni.TCatSubBrick()
>>> tcsb.inputs.in_files = [('functional.nii', "'{2..$}'"), ('functional2.nii', "'{2..$}'")]
>>> tcsb.inputs.out_file= 'functional_tcat.nii'
>>> tcsb.inputs.rlt = '+'
>>> tcsb.cmdline
"3dTcat -rlt+ -prefix functional_tcat.nii functional.nii'{2..$}' functional2.nii'{2..$}' "
>>> res = tcsb.run()
Inputs:
[Mandatory]
in_files: (a list of items which are a tuple of the form: (an
existing file name, a unicode string))
List of tuples of file names and subbrick selectors as strings.Don't
forget to protect the single quotes in the subbrick selectorso the
contents are protected from the command line interpreter.
argument: ``%s%s ...``, position: -1
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``
rlt: ('' or '+' or '++')
Remove linear trends in each voxel time series loaded from each
input dataset, SEPARATELY. Option -rlt removes the least squares fit
of 'a+b*t' to each voxel time series. Option -rlt+ adds dataset mean
back in. Option -rlt++ adds overall mean of all dataset timeseries
back in.
argument: ``-rlt%s``, position: 1
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
TStat¶
Wraps the executable command 3dTstat.
Compute voxel-wise statistics using AFNI 3dTstat command
For complete details, see the 3dTstat Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> tstat = afni.TStat()
>>> tstat.inputs.in_file = 'functional.nii'
>>> tstat.inputs.args = '-mean'
>>> tstat.inputs.out_file = 'stats'
>>> tstat.cmdline
'3dTstat -mean -prefix stats functional.nii'
>>> res = tstat.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dTstat
argument: ``%s``, position: -1
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``
mask: (an existing file name)
mask file
argument: ``-mask %s``
options: (a unicode string)
selected statistical output
argument: ``%s``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
To3D¶
Wraps the executable command to3d.
Create a 3D dataset from 2D image files using AFNI to3d command
For complete details, see the to3d Documentation
Examples¶
>>> from nipype.interfaces import afni
>>> to3d = afni.To3D()
>>> to3d.inputs.datatype = 'float'
>>> to3d.inputs.in_folder = '.'
>>> to3d.inputs.out_file = 'dicomdir.nii'
>>> to3d.inputs.filetype = 'anat'
>>> to3d.cmdline
'to3d -datum float -anat -prefix dicomdir.nii ./*.dcm'
>>> res = to3d.run()
Inputs:
[Mandatory]
in_folder: (an existing directory name)
folder with DICOM images to convert
argument: ``%s/*.dcm``, position: -1
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``
filetype: ('spgr' or 'fse' or 'epan' or 'anat' or 'ct' or 'spct' or
'pet' or 'mra' or 'bmap' or 'diff' or 'omri' or 'abuc' or 'fim' or
'fith' or 'fico' or 'fitt' or 'fift' or 'fizt' or 'fict' or 'fibt'
or 'fibn' or 'figt' or 'fipt' or 'fbuc')
type of datafile being converted
argument: ``-%s``
skipoutliers: (a boolean)
skip the outliers check
argument: ``-skip_outliers``
assumemosaic: (a boolean)
assume that Siemens image is mosaic
argument: ``-assume_dicom_mosaic``
datatype: ('short' or 'float' or 'byte' or 'complex')
set output file datatype
argument: ``-datum %s``
funcparams: (a unicode string)
parameters for functional data
argument: ``-time:zt %s alt+z2``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
Undump¶
Wraps the executable command 3dUndump.
3dUndump - Assembles a 3D dataset from an ASCII list of coordinates and (optionally) values.
The input file(s) are ASCII files, with one voxel specification per line. A voxel specification is 3 numbers (-ijk or -xyz coordinates), with an optional 4th number giving the voxel value. For example:
1 2 3 3 2 1 5 5.3 6.2 3.7 // this line illustrates a comment
The first line puts a voxel (with value given by ‘-dval’) at point (1,2,3). The second line puts a voxel (with value 5) at point (3,2,1). The third line puts a voxel (with value given by ‘-dval’) at point (5.3,6.2,3.7). If -ijk is in effect, and fractional coordinates are given, they will be rounded to the nearest integers; for example, the third line would be equivalent to (i,j,k) = (5,6,4).
For complete details, see the 3dUndump Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> unndump = afni.Undump()
>>> unndump.inputs.in_file = 'structural.nii'
>>> unndump.inputs.out_file = 'structural_undumped.nii'
>>> unndump.cmdline
'3dUndump -prefix structural_undumped.nii -master structural.nii'
>>> res = unndump.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dUndump, whose geometry will determinethe geometry of
the output
argument: ``-master %s``, position: -1
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``
mask_file: (a file name)
mask image file name. Only voxels that are nonzero in the mask can
be set.
argument: ``-mask %s``
datatype: ('short' or 'float' or 'byte')
set output file datatype
argument: ``-datum %s``
default_value: (a float)
default value stored in each input voxel that does not have a value
supplied in the input file
argument: ``-dval %f``
fill_value: (a float)
value, used for each voxel in the output dataset that is NOT listed
in the input file
argument: ``-fval %f``
coordinates_specification: ('ijk' or 'xyz')
Coordinates in the input file as index triples (i, j, k) or spatial
coordinates (x, y, z) in mm
argument: ``-%s``
srad: (a float)
radius in mm of the sphere that will be filled about each input
(x,y,z) or (i,j,k) voxel. If the radius is not given, or is 0, then
each input data line sets the value in only one voxel.
argument: ``-srad %f``
orient: (a tuple of the form: ('R' or 'L', 'A' or 'P', 'I' or 'S'))
Specifies the coordinate order used by -xyz. The code must be 3
letters, one each from the pairs {R,L} {A,P} {I,S}. The first letter
gives the orientation of the x-axis, the second the orientation of
the y-axis, the third the z-axis: R = right-to-left L = left-to-
right A = anterior-to-posterior P = posterior-to-anterior I =
inferior-to-superior S = superior-to-inferior If -orient isn't used,
then the coordinate order of the -master (in_file) dataset is used
to interpret (x,y,z) inputs.
argument: ``-orient %s``
head_only: (a boolean)
create only the .HEAD file which gets exploited by the AFNI matlab
library function New_HEAD.m
argument: ``-head_only``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
assembled file
References:¶
None None
Unifize¶
Wraps the executable command 3dUnifize.
3dUnifize - for uniformizing image intensity
- The input dataset is supposed to be a T1-weighted volume, possibly already skull-stripped (e.g., via 3dSkullStrip). However, this program can be a useful step to take BEFORE 3dSkullStrip, since the latter program can fail if the input volume is strongly shaded – 3dUnifize will (mostly) remove such shading artifacts.
- The output dataset has the white matter (WM) intensity approximately uniformized across space, and scaled to peak at about 1000.
- The output dataset is always stored in float format!
- If the input dataset has more than 1 sub-brick, only sub-brick #0 will be processed!
- Want to correct EPI datasets for nonuniformity? You can try the new and experimental [Mar 2017] ‘-EPI’ option.
- The principal motive for this program is for use in an image registration script, and it may or may not be useful otherwise.
- This program replaces the older (and very different) 3dUniformize, which is no longer maintained and may sublimate at any moment. (In other words, we do not recommend the use of 3dUniformize.)
For complete details, see the 3dUnifize Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> unifize = afni.Unifize()
>>> unifize.inputs.in_file = 'structural.nii'
>>> unifize.inputs.out_file = 'structural_unifized.nii'
>>> unifize.cmdline
'3dUnifize -prefix structural_unifized.nii -input structural.nii'
>>> res = unifize.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dUnifize
argument: ``-input %s``, position: -1
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``
t2: (a boolean)
Treat the input as if it were T2-weighted, rather than T1-weighted.
This processing is done simply by inverting the image contrast,
processing it as if that result were T1-weighted, and then re-
inverting the results counts of voxel overlap, i.e., each voxel will
contain the number of masks that it is set in.
argument: ``-T2``
gm: (a boolean)
Also scale to unifize 'gray matter' = lower intensity voxels (to aid
in registering images from different scanners).
argument: ``-GM``
urad: (a float)
Sets the radius (in voxels) of the ball used for the sneaky trick.
Default value is 18.3, and should be changed proportionally if the
dataset voxel size differs significantly from 1 mm.
argument: ``-Urad %s``
scale_file: (a file name)
output file name to save the scale factor used at each voxel
argument: ``-ssave %s``
no_duplo: (a boolean)
Do NOT use the 'duplo down' step; this can be useful for lower
resolution datasets.
argument: ``-noduplo``
epi: (a boolean)
Assume the input dataset is a T2 (or T2*) weighted EPI time series.
After computing the scaling, apply it to ALL volumes (TRs) in the
input dataset. That is, a given voxel will be scaled by the same
factor at each TR. This option also implies '-noduplo' and
'-T2'.This option turns off '-GM' if you turned it on.
argument: ``-EPI``
mutually_exclusive: gm
requires: no_duplo, t2
rbt: (a tuple of the form: (a float, a float, a float))
Option for AFNI experts only.Specify the 3 parameters for the
algorithm:
R = radius; same as given by option '-Urad', [default=18.3]
b = bottom percentile of normalizing data range, [default=70.0]
r = top percentile of normalizing data range, [default=80.0]
argument: ``-rbt %f %f %f``
t2_up: (a float)
Option for AFNI experts only.Set the upper percentile point used for
T2-T1 inversion. Allowed to be anything between 90 and 100
(inclusive), with default to 98.5 (for no good reason).
argument: ``-T2up %f``
cl_frac: (a float)
Option for AFNI experts only.Set the automask 'clip level fraction'.
Must be between 0.1 and 0.9. A small fraction means to make the
initial threshold for clipping (a la 3dClipLevel) smaller, which
will tend to make the mask larger. [default=0.1]
argument: ``-clfrac %f``
quiet: (a boolean)
Don't print the progress messages.
argument: ``-quiet``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
scale_file: (a file name)
scale factor file
out_file: (an existing file name)
unifized file
References:¶
None None
ZCutUp¶
Wraps the executable command 3dZcutup.
Cut z-slices from a volume using AFNI 3dZcutup command
For complete details, see the 3dZcutup Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> zcutup = afni.ZCutUp()
>>> zcutup.inputs.in_file = 'functional.nii'
>>> zcutup.inputs.out_file = 'functional_zcutup.nii'
>>> zcutup.inputs.keep= '0 10'
>>> zcutup.cmdline
'3dZcutup -keep 0 10 -prefix functional_zcutup.nii functional.nii'
>>> res = zcutup.run()
Inputs:
[Mandatory]
in_file: (an existing file name)
input file to 3dZcutup
argument: ``%s``, position: -1
[Optional]
out_file: (a file name)
output image file name
argument: ``-prefix %s``
keep: (a unicode string)
slice range to keep in output
argument: ``-keep %s``
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
Zcat¶
Wraps the executable command 3dZcat.
Copies an image of one type to an image of the same or different type using 3dZcat command
For complete details, see the 3dZcat Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> zcat = afni.Zcat()
>>> zcat.inputs.in_files = ['functional2.nii', 'functional3.nii']
>>> zcat.inputs.out_file = 'cat_functional.nii'
>>> zcat.cmdline
'3dZcat -prefix cat_functional.nii functional2.nii functional3.nii'
>>> res = zcat.run()
Inputs:
[Mandatory]
in_files: (a list of items which are an existing file name)
argument: ``%s``, position: -1
[Optional]
out_file: (a file name)
output dataset prefix name (default 'zcat')
argument: ``-prefix %s``
datum: ('byte' or 'short' or 'float')
specify data type for output. Valid types are 'byte', 'short' and
'float'.
argument: ``-datum %s``
verb: (a boolean)
print out some verbositiness as the program proceeds.
argument: ``-verb``
fscale: (a boolean)
Force scaling of the output to the maximum integer range. This only
has effect if the output datum is byte or short (either forced or
defaulted). This option is sometimes necessary to eliminate
unpleasant truncation artifacts.
argument: ``-fscale``
mutually_exclusive: nscale
nscale: (a boolean)
Don't do any scaling on output to byte or short datasets. This may
be especially useful when operating on mask datasets whose output
values are only 0's and 1's.
argument: ``-nscale``
mutually_exclusive: fscale
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None
Zeropad¶
Wraps the executable command 3dZeropad.
Adds planes of zeros to a dataset (i.e., pads it out).
For complete details, see the 3dZeropad Documentation.
Examples¶
>>> from nipype.interfaces import afni
>>> zeropad = afni.Zeropad()
>>> zeropad.inputs.in_files = 'functional.nii'
>>> zeropad.inputs.out_file = 'pad_functional.nii'
>>> zeropad.inputs.I = 10
>>> zeropad.inputs.S = 10
>>> zeropad.inputs.A = 10
>>> zeropad.inputs.P = 10
>>> zeropad.inputs.R = 10
>>> zeropad.inputs.L = 10
>>> zeropad.cmdline
'3dZeropad -A 10 -I 10 -L 10 -P 10 -R 10 -S 10 -prefix pad_functional.nii functional.nii'
>>> res = zeropad.run()
Inputs:
[Mandatory]
in_files: (an existing file name)
input dataset
argument: ``%s``, position: -1
[Optional]
out_file: (a file name)
output dataset prefix name (default 'zeropad')
argument: ``-prefix %s``
I: (an integer (int or long))
adds 'n' planes of zero at the Inferior edge
argument: ``-I %i``
mutually_exclusive: master
S: (an integer (int or long))
adds 'n' planes of zero at the Superior edge
argument: ``-S %i``
mutually_exclusive: master
A: (an integer (int or long))
adds 'n' planes of zero at the Anterior edge
argument: ``-A %i``
mutually_exclusive: master
P: (an integer (int or long))
adds 'n' planes of zero at the Posterior edge
argument: ``-P %i``
mutually_exclusive: master
L: (an integer (int or long))
adds 'n' planes of zero at the Left edge
argument: ``-L %i``
mutually_exclusive: master
R: (an integer (int or long))
adds 'n' planes of zero at the Right edge
argument: ``-R %i``
mutually_exclusive: master
z: (an integer (int or long))
adds 'n' planes of zero on EACH of the dataset z-axis (slice-
direction) faces
argument: ``-z %i``
mutually_exclusive: master
RL: (an integer (int or long))
specify that planes should be added or cut symmetrically to make the
resulting volume haveN slices in the right-left direction
argument: ``-RL %i``
mutually_exclusive: master
AP: (an integer (int or long))
specify that planes should be added or cut symmetrically to make the
resulting volume haveN slices in the anterior-posterior direction
argument: ``-AP %i``
mutually_exclusive: master
IS: (an integer (int or long))
specify that planes should be added or cut symmetrically to make the
resulting volume haveN slices in the inferior-superior direction
argument: ``-IS %i``
mutually_exclusive: master
mm: (a boolean)
pad counts 'n' are in mm instead of slices, where each 'n' is an
integer and at least 'n' mm of slices will be added/removed; e.g., n
= 3 and slice thickness = 2.5 mm ==> 2 slices added
argument: ``-mm``
mutually_exclusive: master
master: (a file name)
match the volume described in dataset 'mset', where mset must have
the same orientation and grid spacing as dataset to be padded. the
goal of -master is to make the output dataset from 3dZeropad match
the spatial 'extents' of mset by adding or subtracting slices as
needed. You can't use -I,-S,..., or -mm with -master
argument: ``-master %s``
mutually_exclusive: I, S, A, P, L, R, z, RL, AP, IS, mm
num_threads: (an integer (int or long), nipype default value: 1)
set number of threads
outputtype: ('NIFTI' or 'AFNI' or 'NIFTI_GZ')
AFNI output filetype
args: (a unicode string)
Additional parameters to the command
argument: ``%s``
environ: (a dictionary with keys which are a bytes or None or a value
of class 'str' and with values which are a bytes or None or a
value of class 'str', nipype default value: {})
Environment variables
Outputs:
out_file: (an existing file name)
output file
References:¶
None None